Tom Goddard
January 26, 2017
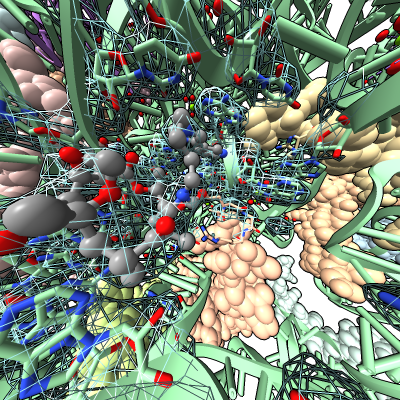
Virtual reality demonstration showing antibiotic bound to E coli ribosome (PDB 4v7s) for reception after Byer's Award Lecture given by Danica Fujimori on "Unlocking the Mystery of Antibiotic Resistance", Tuesday January 31, 4:30 PM. Reception is in Genentech Hall atrium where HTC Vive demo will be set up for public viewing.
Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating enzyme.
Nucleic Acids Res. 2016 Oct 14;44(18):8897-8907. Epub 2016 Aug 5.
Stojković V1, Noda-Garcia L2, Tawfik DS3, Fujimori DG4.
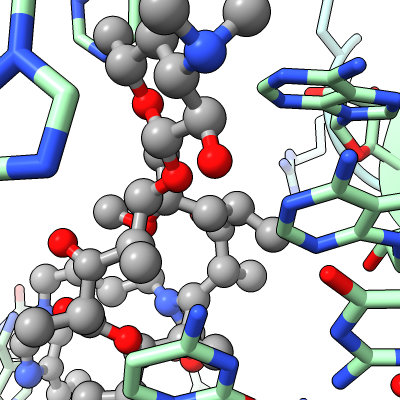
Work describes antibiotic resistance to antibiotics that bind to the peptidyl transferase center (PTC) of bacterial ribosomes caused by mutations which suppress an endogenous post-tranlational methylation of 23S ribosomal RNA base A2503 at C2. Other antibiotic resistance is conferred by methylation of C8 of this base. The C8 atom right next to the antibiotic while the C2 atom is on the opposite side some distance from the antibiotic but likely C2 methylation effects positioning of the base.
ChimeraX command file scene1.cxc to setup ribosome scene. To limit scene to zone around antibiotic for manipulating with Vive hand controllers run scene2.cxc after scene1.cxc. To reset from zone scene back to full ribosome scene use scene3.cxc.
This very large scene can be viewed without stuttering using a Geforce GTX 1080. May stutter on slower graphics cards.
Show two VR views, one immersive at near the buried antibiotic binding site. Don't use hand controllers since moving the scene with structure all around is highly nauseating. Second scene shows just antibiotic and nearby residues allowing hand controllers for zooming and moving model in front of viewer. scene.
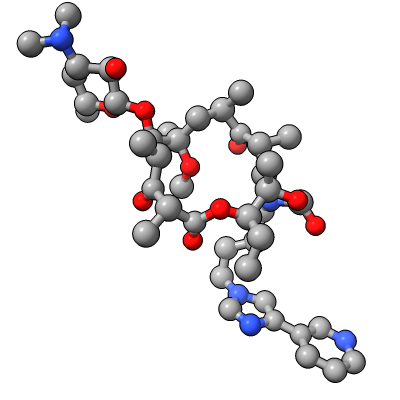
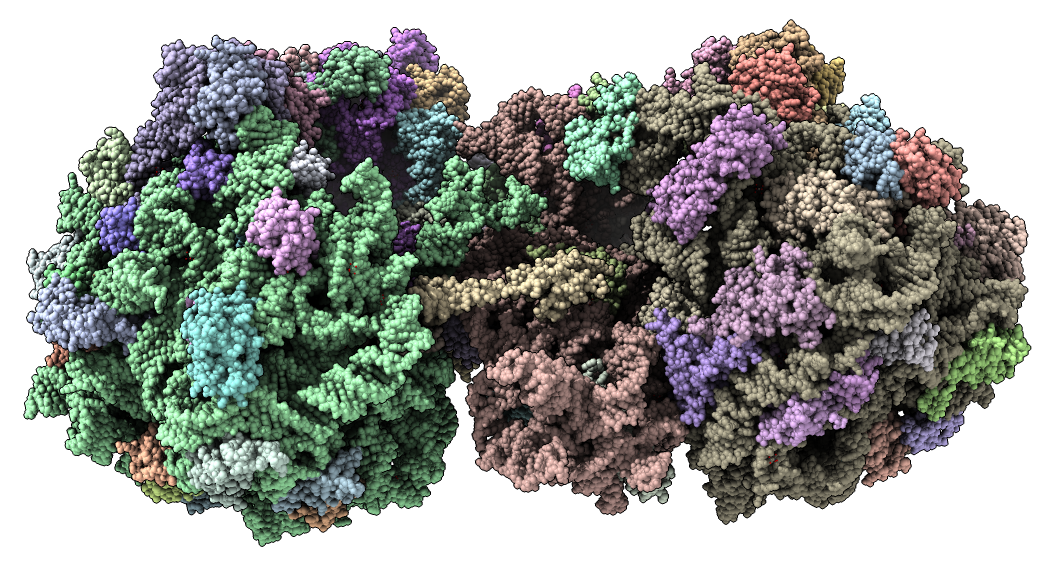
Show E. coli ribosome with telithromycin antibiotic bound (residue name TEL), PDB 4v7s, a 2010 structure from the Jamie Cate lab at UC Berkeley. Structure contains two copies of full ribosome in crystal asymmetric unit.
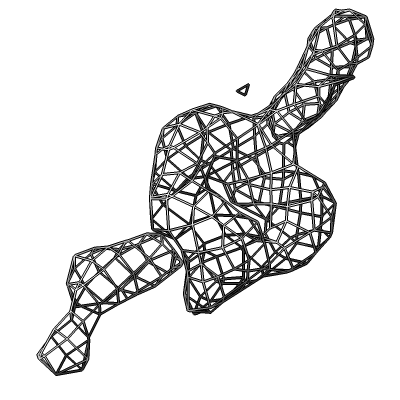
Also show x-ray density (3.25 Angstrom) mesh from Electron Density server.

| 
| 
| 
|
| Antibiotic binding site | Ribosomal RNA in rungs style where each nucleotide shown as a cylinder | Antibiotic telithromycin | Ribosomal sidechains near antibiotic shown in stick style |
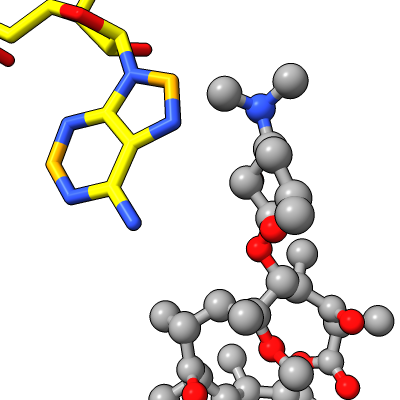
| 
| 
| 
|
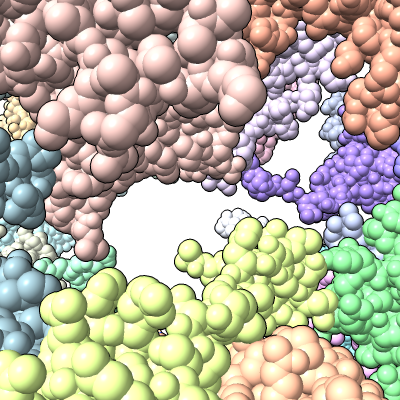
| Methylation at nucleotide A2503 (yellow) C2 and C8 atoms (orange) related to antibiotic resistance | X-ray crystallography density | Ribosomal proteins shown as spheres | Magnesium ions with metal coordination bonds |
Ribosome-targeting antibiotics and mechanisms of bacterial resistance
Daniel N. Wilson
Nature Reviews Microbiology 12, 35–48 (2014) doi:10.1038/nrmicro3155
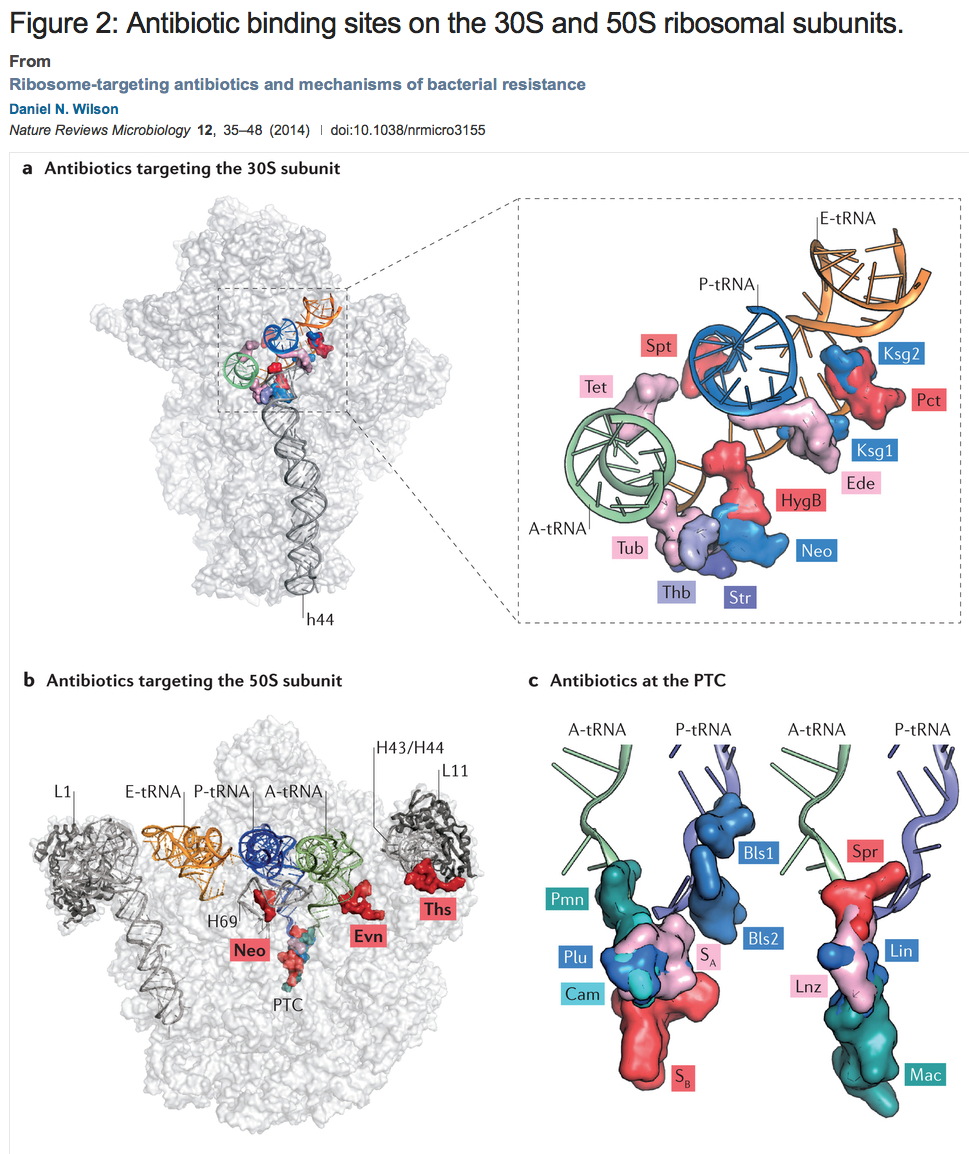
Figure: a | Overview and enlargement of antibiotic binding sites along the mRNA binding channel of the 30S subunit, including tetracycline (Tet; Protein Data Bank (PDB) accession 4G5K–N)18, spectinomycin (Spt; PDB accession 2QOU–X)139, kasugamycin (Ksg1 and Ksg2; PDB accession 2HHH)140, pactamycin (Pct; PDB accession 1HNX)13, edeine (Ede; PDB accession 1I95)14, hygromycin B (HygB; PDB accession 3DF1–4)141, neomycin (Neo; PDB accession 4GAQ/R/S/U)25, streptomycin (Str; PDB accession 1FJG)142, thermorubin (Thb; PDB accession 3UXQ–T)30 and tuberactinomycins (Tub; PDB accession 3KNH–K)29. The A-site tRNA (green), P-site tRNA (blue), E-site tRNA (orange) and h44 are highlighted for reference. b | Overview of the binding sites of neomycin (Neo; PDB accession 4GAQ/R/S/U)25, evernimicin (Evn)10 and thiostrepton (Ths; PDB accession 3CF5)81 on the 50S subunit. The A-site tRNA (green), P-site tRNA (blue), E-site tRNA (orange), H43/H44, H69, peptidyl-transferase centre (PTC) and the L1 and L11 stalks are highlighted for reference. c | Enlargement of the binding sites of blasticidin S (Bls1 and Bls2; PDB accession 1KC8)47, sparsomycin (Spr; PDB accession 1M90)143, lincomycin (Lin; PDB accession 3OFX/Y/Z/0)41, linezolid (Lnz; PDB accession 3DLL)53, macrolides (Mac; PDB accession 1K9M)43, puromycin (Pmn; PDB accession 1M90)143, pleuromutilins (Plu; PDB accession 1XBP)144, chloramphenicol (Cam; PDB accession 3OFA–D)41 and streptogramins A and B (SA and SB; PDB accession 1SM1)68 relative to the A-site and P-site tRNAs.