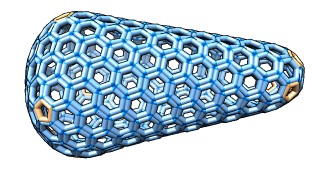
Cage Builder 
Cage Builder creates polyhedral cages composed of hexagons, pentagons,
squares and other polygons. The cages can be decorated with atomic models
to create oligomeric molecular assemblies
using the sym command
(see examples).
Cages are created as
marker sets.
See also:
Icosahedron Surface,
hkcage,
meshmol
There are several ways to start
Cage Builder, a tool in the
Higher-Order Structure category.
-
Attach polygons with [ 3 ][ 4 ][ 5 ][ 6 ][ 7 ] sides
- clicking a number attaches a new polygon of the specified shape
to each currently selected
polygon edge, or if no such edges exist, creates a single new polygon.
All polygons are part of a single Chimera
marker model
named Cage. The attached edges are automatically joined.
Joined edges are shown in lighter colors than unjoined edges.
Clicking Minimize improves the alignment of joined edges.
This is typically done after some rounds of new polgyon attachment.
Clicking Delete removes entire polygons that contain any
selected edge or vertex.
Close simply dismisses the dialog, and
Help opens this manual page in a browser window.
Clicking Options reveals additional settings
(clicking the close button  on the right
hides them again):
on the right
hides them again):
- [ Join ] or [ Unjoin ] selected polygon edges
- clicking Join when exactly two edges are
selected will join those edges.
The two edges should belong to different polygons, and a polygon will be
relocated as needed to attach and join the edges.
Their previous join partners, if any, will become unjoined;
an edge can only be joined to one other edge at a time.
Clicking Join with fewer or more than two edges selected has no effect.
Unjoin will unjoin all joined pairs with one or both edges selected.
- Join polygons so that [N] edges meet at a vertex
(default on)
- whether to automatically join additional edges during new polygon attachment
or when Join is used, with the constraint that N edges
(default 3) should meet at each vertex.
For example, if a lone pentagon has all of its edges selected
and the 6 button is clicked, not only will a new hexagon
be attached (adjacent edges joined) to each side of the pentagon,
but also each hexagon will be attached its neighboring hexagons.
Polygon positions may be adjusted to accommodate joining.
If this option is off, only the edges of new
polygon attachment will be joined automatically.
- [ Scale ] cage by [f]
- clicking Scale or changing the value and pressing Enter (return)
resizes all polygons and moves them radially
from the geometric center of the cage model by a factor f
(default 2)
- [ Set ] edge length [e] thickness [t]
• inset [i] (default on)
- clicking Set or changing a value and pressing Enter (return)
updates all polygons in the cage model to the specified
edge length (default 1.0) and stick thickness (default 0.2)
in physical units, typically Å.
With insetting on, the edge lengths are reduced by the inset value
(default 0.1)
so that the edges of attached polygons will not overlap completely.
For well-aligned joined edges, using an inset equal to half the thickness
makes the sticks representing the edges lie side by side.
With insetting off, the polygons are shown at the nominal size
with attached edges right on top of each other
(to the extent allowed by geometric contraints).
The length, thickness, and inset settings will also be used
for subsequently created polygons.
- [ Expand ] cage by [g] times edge length
- clicking Expand or changing the value and pressing Enter (return)
fattens the cage by moving each polygon outward
along its normal by a factor g (default 1) times edge length,
keeping polygon sizes the same.
Subsequently clicking Minimize will bring the polygons
back together, often leaving the whole cage with a rounder shape.
This option has rather limited usefulness.
- Create [ Mesh ] model from cage polygons, color
(a color well,
by default a dark gray)
- clicking Mesh generates a new
marker model
from the existing polygons, in which attached edges are replaced by
a single edge and attached vertices are replaced by a single vertex.
The mesh model cannot be edited or resized with the Cage Builder dialog;
all editing should be done with the polygon cage model.
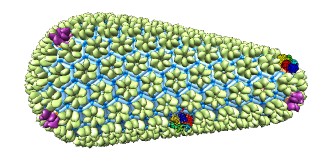
Copies of molecular models can be placed on each polygon using the
sym command.
For example, one might want to put a single copy of the structure
at each pentagon (ignoring any other types of polygons).
First, the molecular model would need to be positioned as desired relative
to one of the pentagons in the cage model.
Models can be positioned relative to one another “manually” by
activating/deactivating
them for motion and using the mouse.
Then, if the molecule model is #1 and the cage
model is #0, the following would place a copy
of the molecule model at an equivalent position relative to each pentagon:
Command:
sym #1 group #0,p5
The copies can be removed with
~sym.
Another possibility is to place multiple copies per polygon. For example,
after a molecule model #1 has been positioned relative to (1/6) of a hexagon
in cage model #0, the following would use C6 symmetry about the
hexagon center to place six copies per hexagon:
Command:
sym
#1 group #0,pn6 surface true update true
The additional arguments in the second example specify creating low-resolution
multiscale surfaces
instead of atomic copies (for faster rendering)
and updating their positions when the original copy of the molecular model
is moved relative to the cage.
Currently there is no good way to refine the molecular positions
so that interfaces between molecules are physically realistic.
UCSF Computer Graphics Laboratory / July 2011
 on the right
hides them again):
on the right
hides them again):
