

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Current Apps (3.x)
News
October 22, 2018
Cytoscape v3.7 has been released. See the release notes for what's new.
March 27, 2017
Cytoscape v3.5 has been released. Releases at the 2.x level are no longer supported.
August 25, 2011
Cytoscape 2.8.2 has been released! This release provides a number of bug fixes, usability improvements, and a significantly improved memory detection and allocation process. Check it out at http://www.cytoscape.org.
(Previous news...)Upcoming Events

The Resource for Biocomputing, Visualization, and Informatics (RBVI) focuses on creating innovative computational and visualization-based data analysis methods and algorithms, implementing them as professional-quality, easy-to-use software tools, and applying these tools to solve a wide range of biomedical research problems. Our historical emphasis has been on developing tools for structural biology, such as the broadly used molecular modeling package, UCSF Chimera.
Recognizing the importance of biological context in understanding the sequence→structure→function triad, we are also developing tools for the analysis and visualization of networks with Cytoscape, a widely used open-source tool. Cytoscape provides an excellent “app” mechanism that we have used to add functionality, and we also participate in the development of the Cytoscape core.
Highlighted App
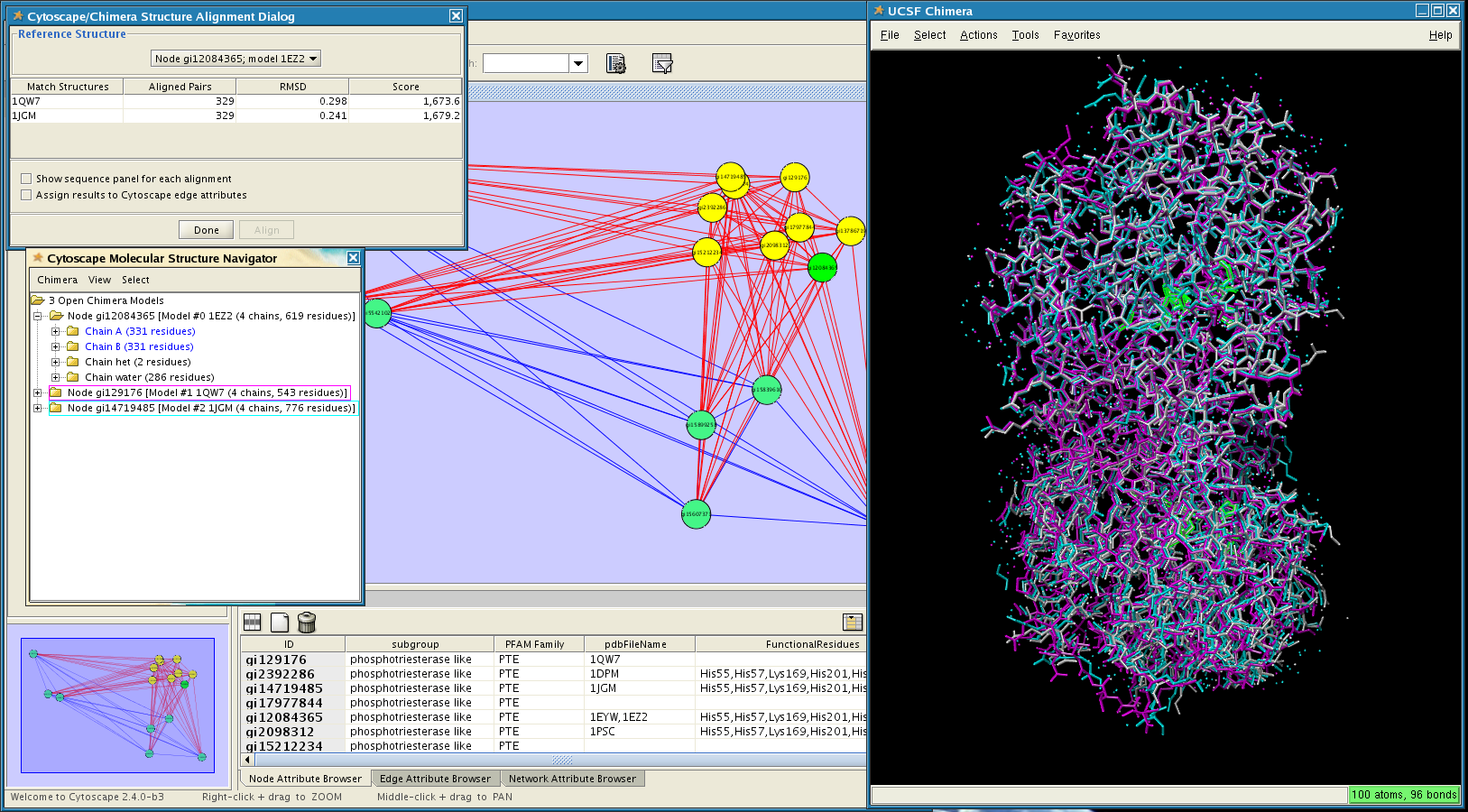
In this screenshot the PFAM family PTE (phosphotriesterase) has been opened in Cytoscape. Three of the available structures have been opened in UCSF Chimera and aligned using Chimera's structural alignment tools. The functional residues of one of the structures (pdb: 1EZ2) have also been selected (green highlights in Chimera).
(More apps...)About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.