 A PDB file being annotated
in an interactive molecule viewer
A PDB file being annotated
in an interactive molecule viewer
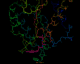
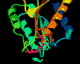 The annotated PDB file
in a ribbon representation
The annotated PDB file
in a ribbon representation
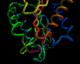 The same annotated PDB file
in a tube representation
The same annotated PDB file
in a tube representation
 The same annotated PDB file
in a CPK representation
The same annotated PDB file
in a CPK representation
A poster by Gregory S. Couch, Eric F. Pettersen, Conrad C. Huang, and Thomas E. Ferrin
Computer Graphics Laboratory
University of California
San Francisco, California 94143-0446
We have implemented extensions to the Brookhaven Protein Data Bank (PDB) file format for incorporating scene information such as viewing parameters, additional molecular information (e.g., van der Waals radii and atom colors) and user-defined graphics. These extensions were made in conformance with the PDB standard and provide sufficient information to render the scene in various styles such as space-filling images and ribbon diagrams. For the past five years these extensions have been used in the MidasPlus molecular modeling system and have proved both powerful and sufficient for generating complex molecular images. We propose that the extensions to the PDB presented here be adopted by the molecular modeling community for incorporation into visualization programs.
The following four images, generated from the same annotated PDB file, show flavodoxin (colored red at the N terminus to blue at the C terminus) and a flavin mononucleotide (FMN) prosthetic group (colored magenta). Just the backbone atoms of flavodoxin are shown as well as the residues that are 3.5 Ångstroms or less away from FMN. Follow the links below to view a full resolution version of the thumbnail images (i.e., a 24-bit 640x512 TIFF image). The flavodoxin complex is the 3fxn entry from the PDB and the reference for the coordinates is: W. W. Smith, R. M. Burnett, G. D. Darling, M. L. Ludwig, "Structure of the Semiquinone Form of Flavodoxin From Clostridium MP. Extension of 1.8 Angstroms Resolution and some Comparisons with the Oxidized State," J. Mol. Biol., 117(?):195, 1977.
 A PDB file being annotated
in an interactive molecule viewer
A PDB file being annotated
in an interactive molecule viewer
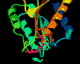 The annotated PDB file
in a ribbon representation
The annotated PDB file
in a ribbon representation
 The same annotated PDB file
in a tube representation
The same annotated PDB file
in a tube representation
 The same annotated PDB file
in a CPK representation
The same annotated PDB file
in a CPK representation
The advantages of our approach to annotating PDB files are:
Notwithstanding the advantages noted above, there are limitations to our method:
Other popular molecular visualization programs use different approaches to combining scene information with PDB files. We briefly discuss a few of these programs below.
Raster3D version 2.0 (with its companion interactive viewer) also separates interactive annotation and rendering into two phases, but abandons the PDB format in favor of a more space-efficient one, thus losing compatibility with existing programs which understand PDB format.
RasMol v2.4 combines an interactive viewer with a renderer. This technique has the advantage that one program is used for both functions, but loses the flexibility of easily incorporating different renderers or being able use filter programs to modify the annotations.
MolScript v1.4 is a PostScript renderer which can embed PDB files into its own input file format. This file format is very flexible, but has the disadvantage of being unintelligible to PDB-conformant programs.
Kinemage is both a file format and an interactive viewer for personal computers. Its chief advantage is that it supports multiple views and bond rotations, but its data files are not in PDB format and are also difficult to edit.
Parts of the preceding poster have been excerpted and paraphrased from a paper published in the Journal of Molecular Graphics 13, 3 (June 1995). A hard-copy reprint of this paper, with color images, is available by sending e-mail to tef@cgl.ucsf.edu.
This work was supported by the NIH National Center for Research Resources, grant P41-RR01081, and by the National Science Foundation, grant IRI-9116999.