
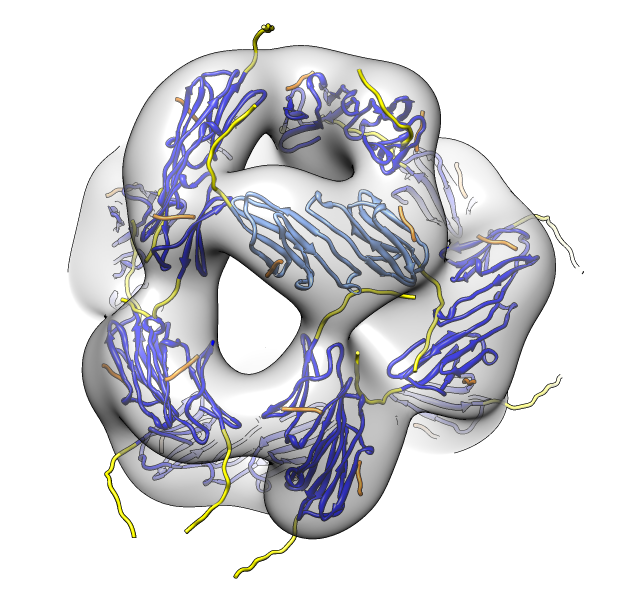
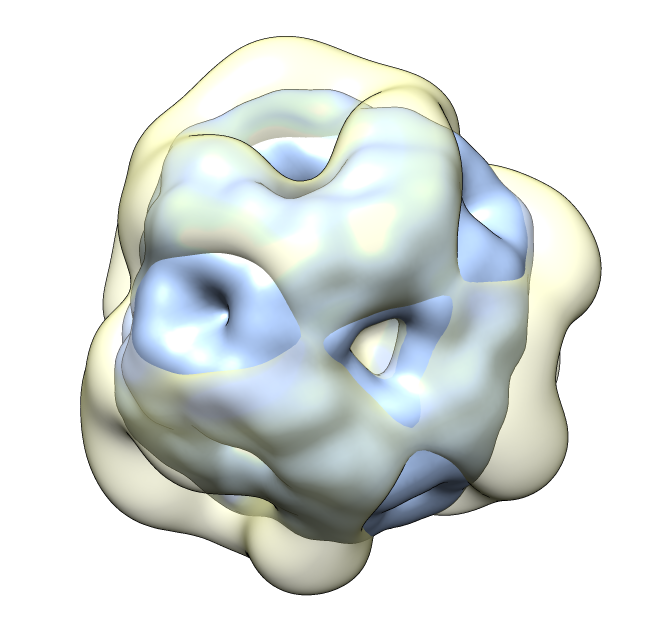
seen by electron microscopy (20 Angstrom resolution).
NCRR Site Visit
November 17, 2011

| 
|
| 24 copies of alpha crystallin form oligomer seen by electron microscopy (20 Angstrom resolution). | Human alpha crystallin is 2 beta sheets. |
|  
| 
| |
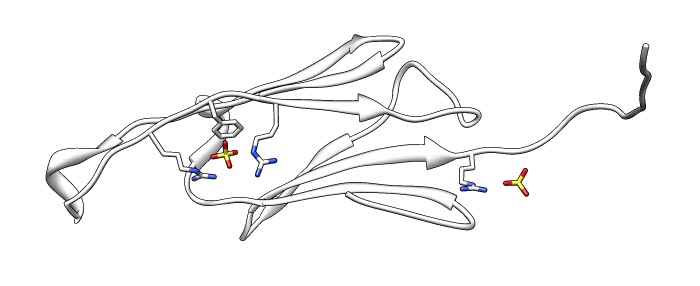
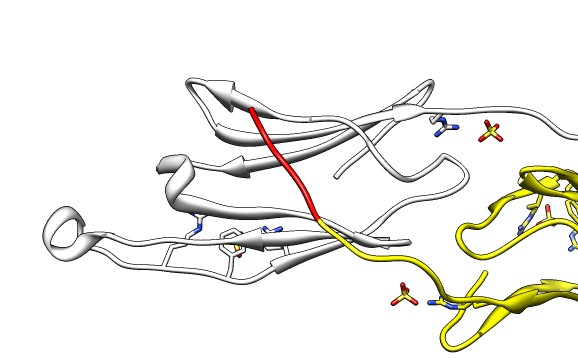
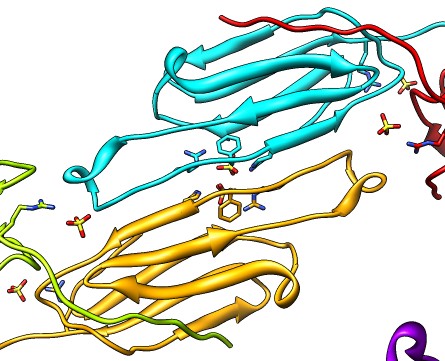
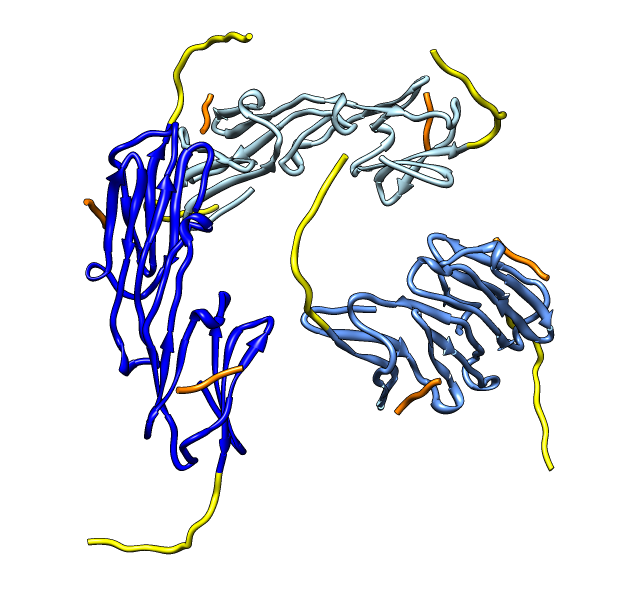
| Various crystal structures show several biological dimerization modes. | C-terminal tail binds to edge of beta-sheet. Binds in 180 degree flipped orientation too. Binding sequence is palindrome. | Side-by-side beta sheet dimer. |
| 
| 
| |
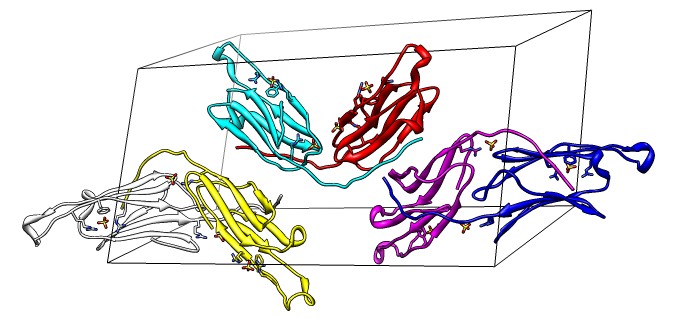
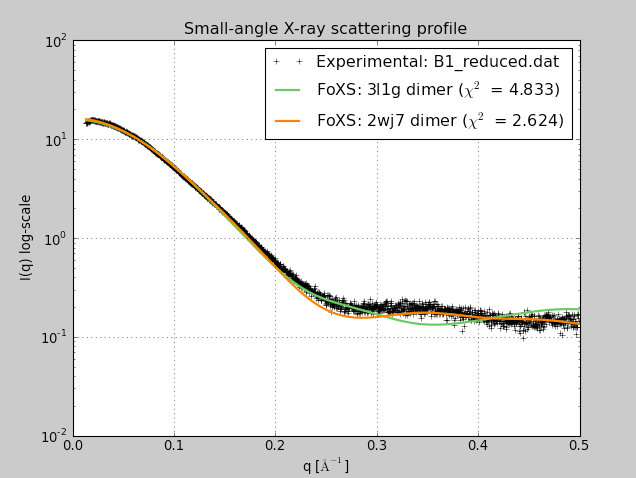
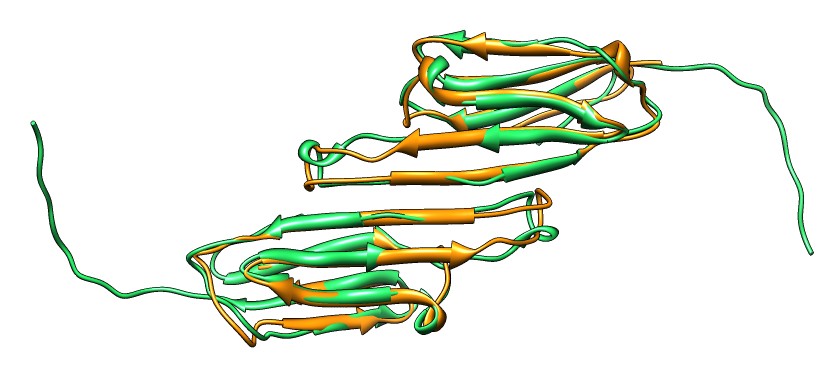
Beta-sheet dimer observed with 3 different registrations in crystals and NMR. Some literature suggests all are present allowing different oligomers to form. | Small-angle x-ray scattering in solution shows only one registration is present at physiological conditions (NMR registration). | Shifted human dimer with C-terminal tails for building 24-mer. |
| 
| 
|
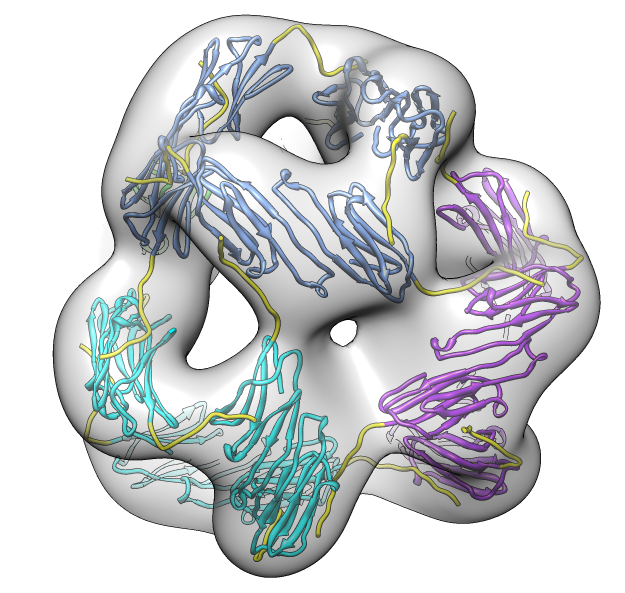
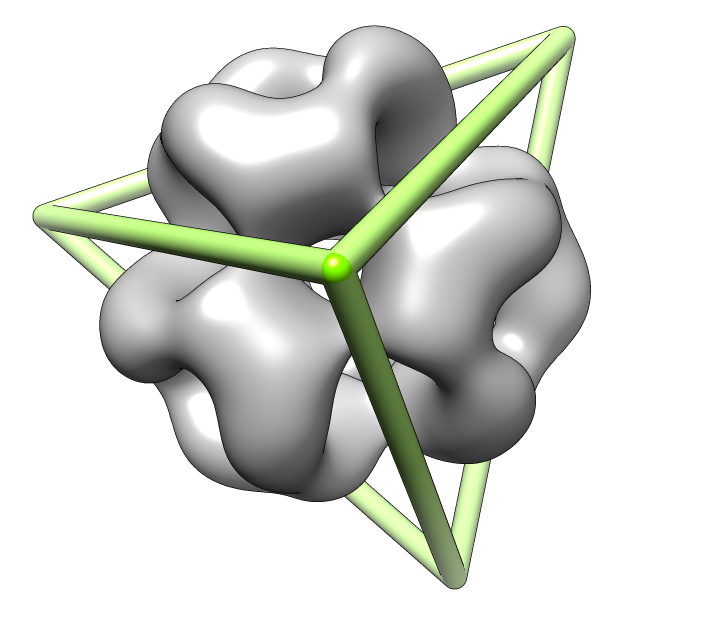
| Deduce map symmetry. | Fit dimer in EM map. Movie. | Symmetry copies of fit dimer. |
| 
| 
| 
|
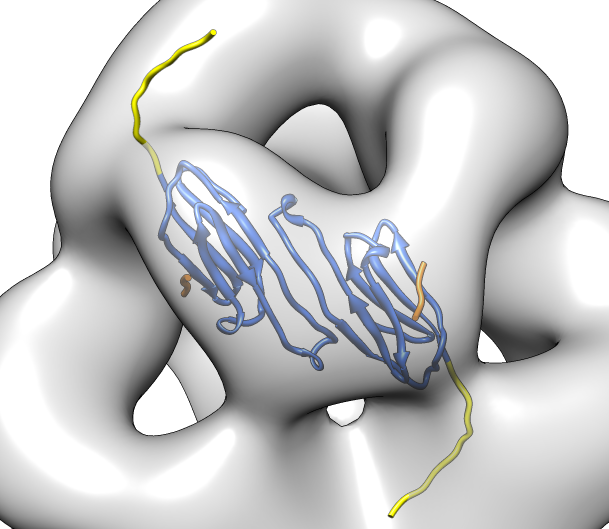
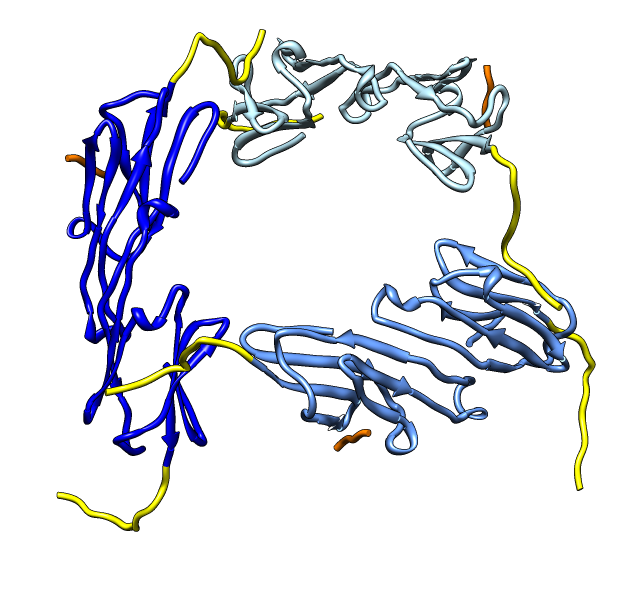
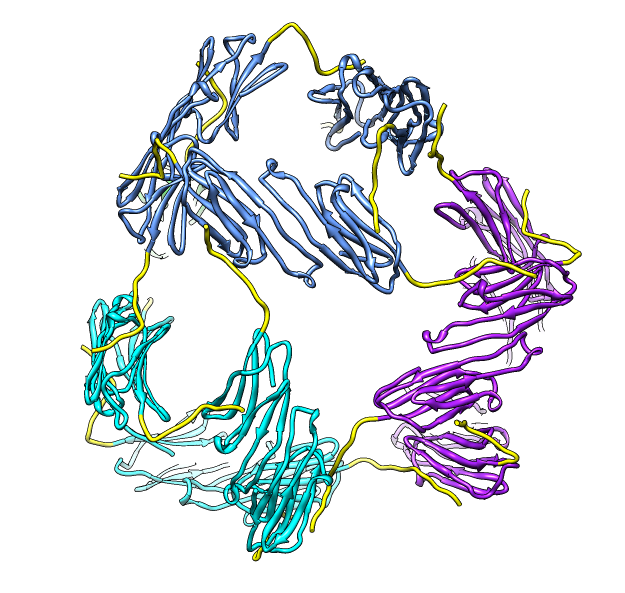
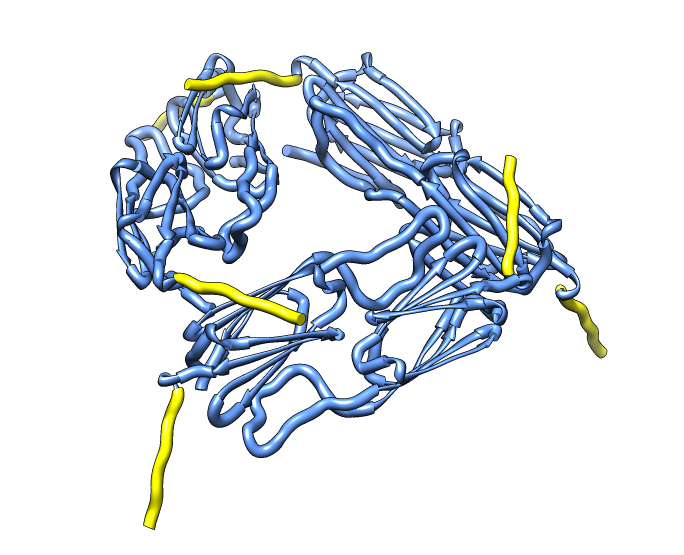
| Hexamer with 6 C-terminal tails. | Moved C-terminal tails to bind hexamer. | Made C-terminal tails connect 4 hexamers. Movie. | Fit of 24-mer model to map. |

| 
| |
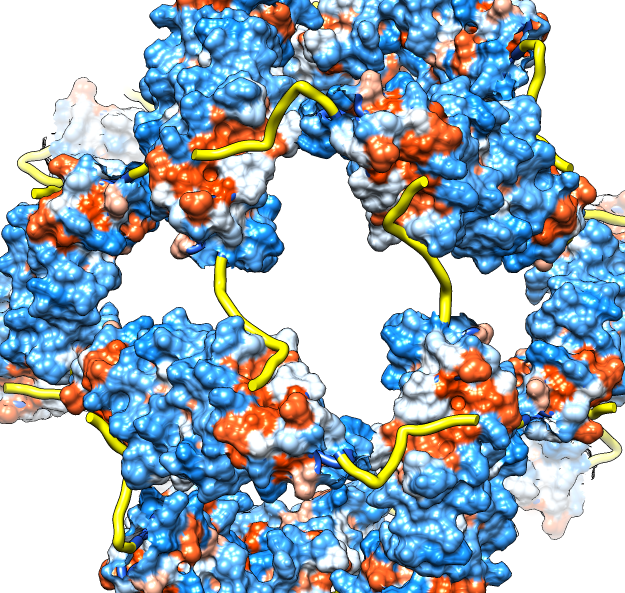
| Molecular surface with orange hydrophobic regions. | C-terminal strands cover hydrophobic grooves. Literature says binding unfolded proteins requires oligomer to disassemble. |
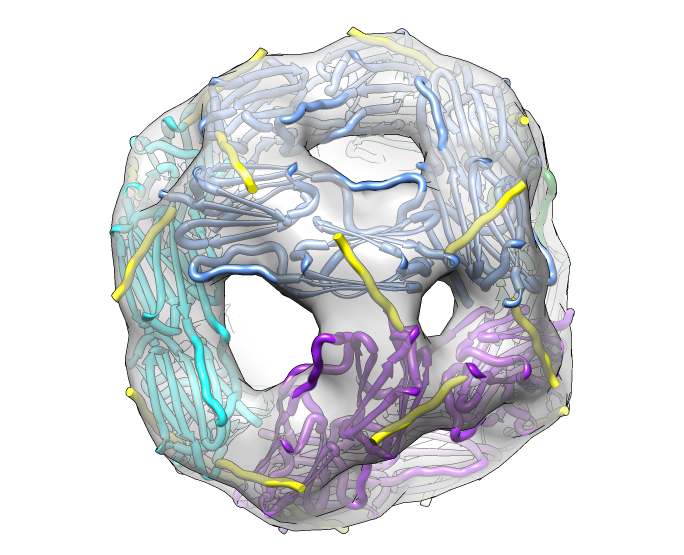
| 
| 
|
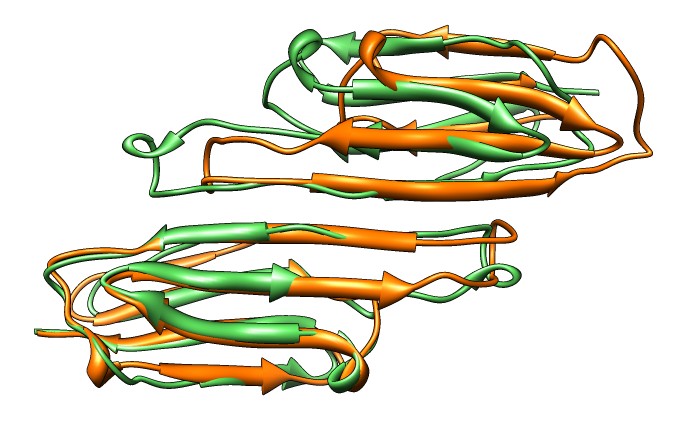
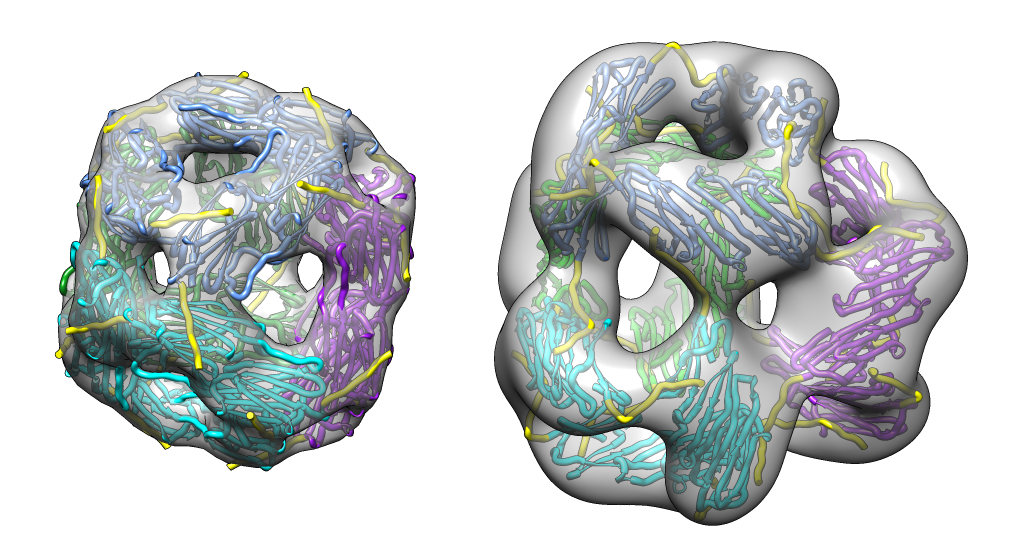
| M jannaschii octahedral small heat shock protein 24-mer (1shs), 20% sequence identity to human alpha-crystallin. | Same C-terminal tail binding pattern as our alpha-crystallin model. | Alpha-crystallin is larger. Two inequivalent of 3-fold axes in alpha-crystallin are equivalent in M jannaschii sHSP. |
| 
| 
|
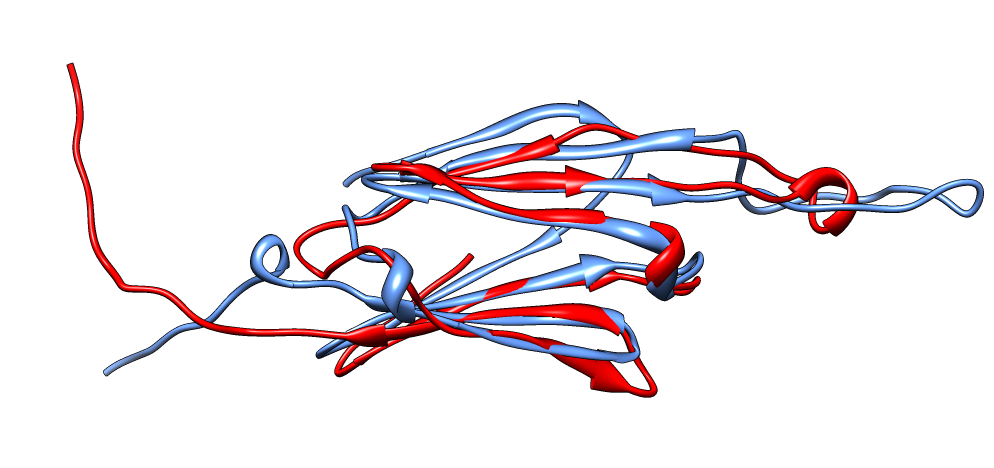
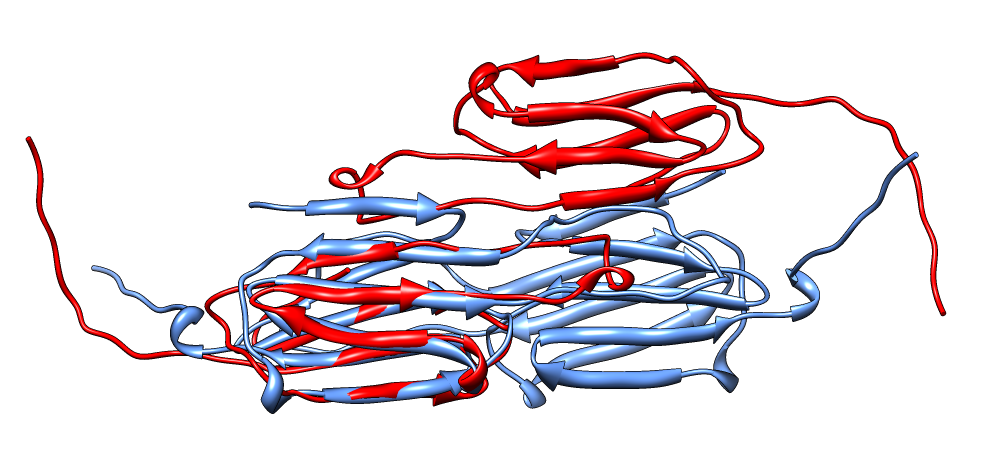
| Aligned alpha-crystallin (red) and sHSP (blue) monomers. | Aligned alpha-crystallin dimer (red), sHSP dimer (blue). Movie. All 12 alpha-crystallin structures show edge-to-edge dimers. None show sHSP type strand swapping. | Comparison of alpha-crystallin EM map and simulated map from sHSP crystal structure. Movie. |
We used 16 Chimera capabilities in this example, only about 1/5 of the available Chimera molecular assembly modeling tools.