HL60 Cell Motion Movies
Tom Goddard
February 15, 2013
Here are some movies of HL60 cells moving as seen by Bessel beam 3-d microscopy.
Movies were made with Chimera. Data from Lillian Fritz-Laylin, Jenny Hsiao,
and Dyche Mullins at UCSF.
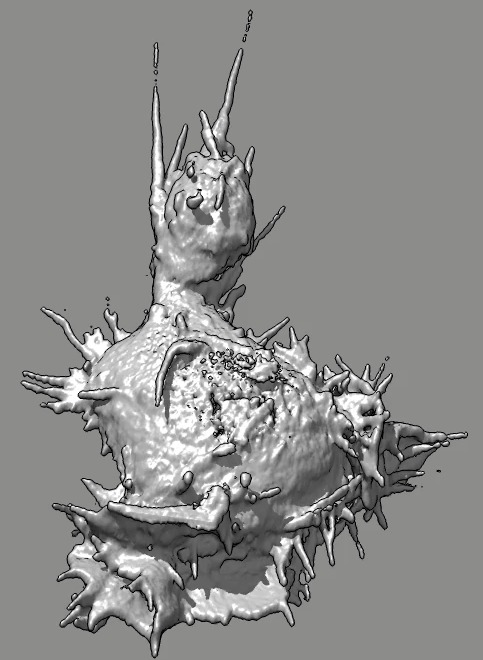
Cell 7 D movie
(11-07-12) top view. Used deconvolved files (*.deconv.am).
Cell goes partly out of view at top of slab. Deconvolution seems
to create high density where cell is cutoff, possibly by including
density values of 100 from outside slab.
|

Cell 7 E movie
(11-07-12) side view. Used deconvolved files (*.deconv.am).
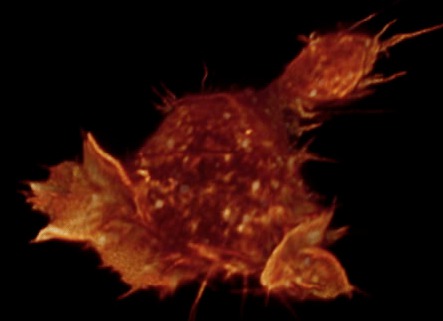
Gray level cell 7 movie
(11-07-12) made by Janelia Farm group, probably with Amira. Chimera
can make this type of movie but I did not try it.
|
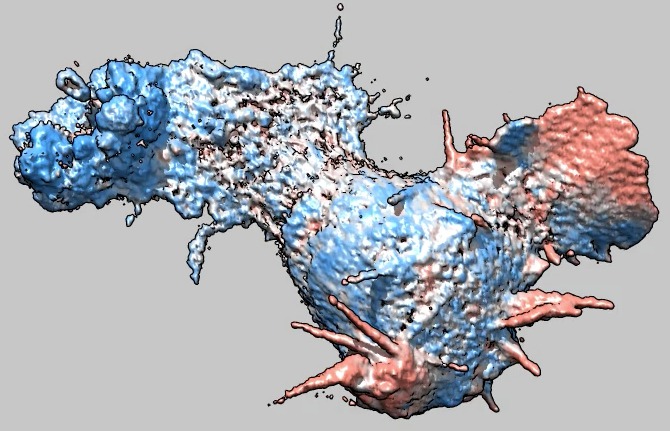
Cell 6 movie
(11-07-12) with moving parts colored red, non-moving blue.
Coloring is done by measuring the correlation between the image at the
current time and the image at the next time over a sliding window 5 by 5 by
5 grid points in size. Colors were blue for correlation 0 (or less), white
for correlation .5, and red (actually salmon) for correlation 1. Colors
were linearly interpolated at other correlation values. Oddly the cell
seems stuck and does not move from its original position.
Used deconvolved files (*.deconv.am).
|
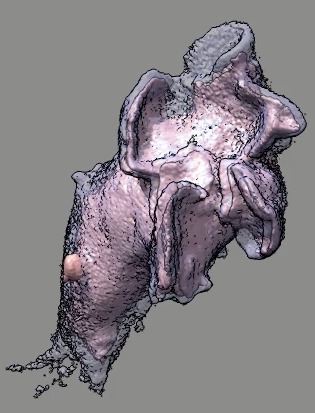
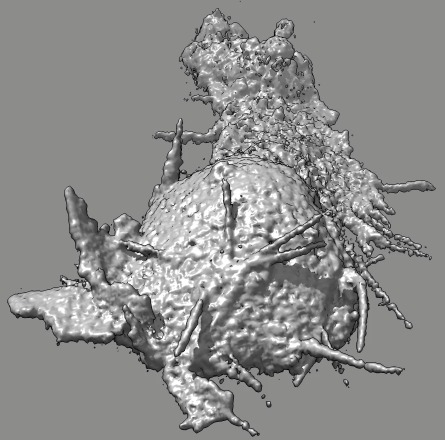
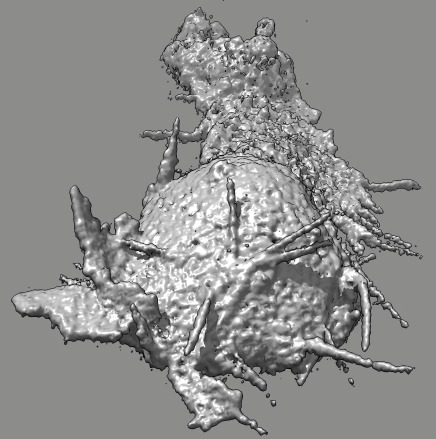
Cell 9 movie
(11-08-12) two channels, channel 1 shown transparent blue,
channel 2 shown opaque pink. Did not use deconvolved files (*.deconv.am)
because they have varying normalization at different times. Used
"translated" .tif files. Filopodia are to some extend smeared out and not
seen in non-deconlved data. Microscope stage moved to keep cell in view.
Cell reverses direction of motion. Cell goes partially out of slab field
of view.
|
|
|
Cell 6 stereo movie
(11-07-12). Left and right eye movies can be played in a stereo
movie player, such as Bino. It can be viewed with
LCD shutter glasses or other stereo viewing techniques such as
cross-eye or wall-eye stereo pairs, or red-blue glasses.
Images in this page show a cross-eye pair (right eye image on left,
left eye image on right).
|
Data Processing
I reduced the size of the data sets so that they can be viewed and played
back in real time. The original data was approximately 250 Mbytes per
frame with 150 to 200 frames so one time series (with one channel) was
is about 40 to 50 Gbytes. At typical disk read speeds of 10-20 seconds per
Gbyte this takes a long time (10 minutes) just to read the data off disk
and does not fit in memory on typical desktop computers.
I reduced the data size about 100-fold using the observation that the
cells only occupied about 1% of the 3d grid (based on an actual measurement
of cell 7 in Chimera). I thresholded the data, setting all values below
a chosen low density level to that level, then compressed the data using
the Chimera "cmap" format based on the HDF5 file format. The thresholding
is needed otherwise the random noise does not allow significant compression.
I also reduced the 32-bit float data values to 16-bit integer to save a factor
of two more in file size. This causes no noticable loss in precision as
the microscope CCD certainly does not exceed this dynamic range.
This reduced the data to typically 0.25 Gbytes which reads quickly and fits
in memory.
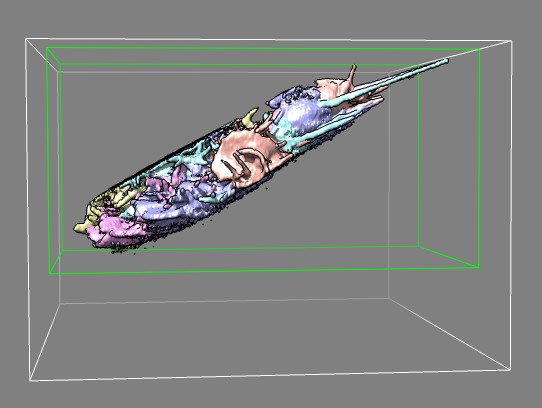 Edge view of several frames of cell 7 (11-07-12) in different colors
showing imaged slab.
Green box was used to crop data to reduce file size for quicker display
in Chimera.
Edge view of several frames of cell 7 (11-07-12) in different colors
showing imaged slab.
Green box was used to crop data to reduce file size for quicker display
in Chimera.
|
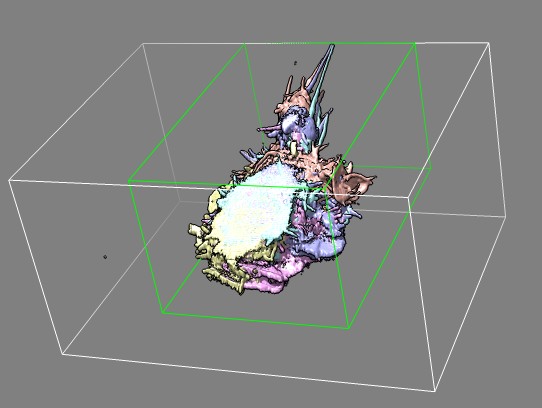 Top view of same cell 7 frames showing that cell is partly out of view
(clipped) at the top.
Top view of same cell 7 frames showing that cell is partly out of view
(clipped) at the top.
|
Here is the Chimera script compress.py
I used to convert the data sets
to compressed cmap format. All time frames are placed in a single .cmap file
for one channel of one time series. Here is an example command to run
this script from a terminal window.
chimera --nogui --nostatus --script "compress.py /Volumes/UCSFdata/JFRC_2012_NOV/11-07-12/cell10_zp4um/*.am cell10.cmap 10,231,0,470,571,180 90 int16"
Converting a data set took about 1 hour on a Mac desktop.
The compressing script allow cropping to a subregion. I inspected several
frames in a time series to see if a cell stayed within a space much smaller
than the data grid. If so I used Chimera subregion selection to find the
reduced bounds. This does not significantly reduce file size, but reduces
the memory and time consumed when individual frames are decompressed in
computer memory. Here's some info on the bounds I chose.
Movie Creation Scripts
Chimera commands to make movies looked like
movie record supersample 3
vseries play #0
wait 150
movie encode ~/Desktop/cell6.mp4 framerate 8 quality high roundtrip true
and were executed by opening a command file such as
record.cmd. The first command starts capturing
images for a movie. The second starts playing the image series, the
third waits for 150 frames to elapse, and the last command writes
an H.264 format movie for playback at 8 frames per second. The
"roundtrip true" option has the movie run the captured sequence forward
and then backward.
movie record supersample 3 ; vseries play #0 ; wait 150 ; movie encode ~/Desktop/cell6.mp4 framerate 8 quality high roundtrip true







 Edge view of several frames of cell 7 (11-07-12) in different colors
showing imaged slab.
Green box was used to crop data to reduce file size for quicker display
in Chimera.
Edge view of several frames of cell 7 (11-07-12) in different colors
showing imaged slab.
Green box was used to crop data to reduce file size for quicker display
in Chimera.
 Top view of same cell 7 frames showing that cell is partly out of view
(clipped) at the top.
Top view of same cell 7 frames showing that cell is partly out of view
(clipped) at the top.