April 11, 2007

| 
|
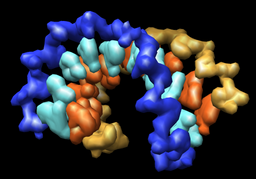
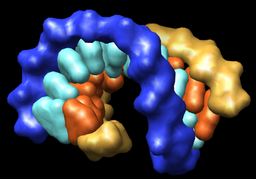
| Multiscale resolution = 1 | Multiscale resolution = 0 |
The reschains.py Python script copies the currently opened molecules putting each nucleic acid base in a separate PDB chain. The nucleic acid backbone and sugars stay in their original chains. This allows the Chimera Multiscale Models tool to create depictions where each backbone and each base is represented by a separate surface.
Open PDB model 100d. Delete waters using menu entries
Select / Chain / water
Actions / Atoms / Delete.
Use menu entry File / Open... to open the reschains.py script. Opening the script runs it, so now each base is in a separate chain.
Show the Multiscale Models dialog with menu entry
Tools / Higher-Order Structure / Multiscale ModelsChange the Resolution setting to 1. Choose Multimer type to be None (near bottom of multiscale dialog). Press the Make models button. This shows the surfaces of backbones and bases.
To use solvent excluded surfaces instead of density style surfaces press the select All button at the top of the Multiscale dialog, change the Resolution to 0, and press Resurface.
The Multiscale Models tool allows coloring the surfaces, changing to other display styles (spheres, ribbons), and changing surface smoothing, resolution, and transparency.
There is a tutorial that explains the Multiscale Models capabilities looking at a virus capsid example. Also the Multiscale page of the Chimera User's Guide gives further information.
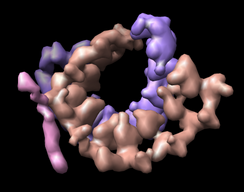
If bases are not placed in separate chains, a single surface including bases and backbone can be made with Multiscale. The image shown used multiscale resolution set to 1. Also I adjusted two other multiscale surfacing parameters by pressing the "..." checkbutton to the right of the Resolution field in the multiscale dialog. I changed "Threshold atom density" to 0.01 to get a fatter surface. And I changed "Smoothing iterations" to 5 to get a smoother surface.
April 11, 2007. Eliminated copying "openedAs" attribute of molecule. This is no longer needed by Multiscale tool in Chimera 1.2251. It caused errors when using the reschains.py script with a scale bar shown or path tracer model opened because those are implemented as molecule objects and do not have the openedAs attribute.
April 5, 2006. Initial version.