Tom Goddard
December 7, 2021

ChimeraX can run AlphaFold on complexes using Google Colab servers. Here is an example of how that is done. This feature is in ChimeraX 1.4 daily builds newer than December 2, 2021 but is not in ChimeraX 1.3.
There is a video that goes through this example.
We will predict the structure of a heterotrimer of photosynthetic reaction center from Rhodobacter sphaeroides. This example is 800 amino acids and took 5 hours to run. We will compare the result to an experimental X-ray structure of this complex, PDB 6Z1J.
This example takes about 5 hours. Leave ChimeraX running. For a minimal dimer example taking 2 hours you can instead use sequences from PDB 7L5M totaling 180 amino acids.
Show the ChimeraX AlphaFold tool with the following menu entry, paste in the 3 amino acid sequences separated by commas, turn on the Is prokaryote option since this complex is from a bacterium, and press the Predict button.
Tools / Structure Prediction / AlphaFold
FDLASLAIYSFWIFLAGLIYYLQTENMREGYPLENEDGTPAANQGPFPLPKPKTFILPHGRGTLTVPGPESEDRPIALARTAVSEGFPHAPTGDPMKDGVGPASW VARRDLPELDGHGHNKIKPMKAAAGFHVSAGKNPIGLPVRGCDLEIAGKVVDIWVDIPEQMARFLEVELKDGSTRLLPMQMVKVQSNRVHVNALSSDLFAGIPTI KSPTEVTLLEEDKICGYVAGGLMYAAPKRKS,
ALLSFERKYRVPGGTLVGGNLFDFWVGPFYVGFFGVATFFFAALGIILIAWSAVLQGTWNPQLISVYPPALEYGLGGAPLAKGGLWQIITICATGAFVSWALREV EICRKLGIGYHIPFAFAFAILAYLTLVLFRPVMMGAWGYAFPYGIWTHLDWVSNTGYTYGNFHYNPAHMIAITFFFTNALALALHGALVLSAANPEKGKEMRTPD HEDTFFRDLVGYSIGTLGIHRLGLLLSLSAVFFSALCMIITGTIWFDQWVDWWQWWVKLPWWANIPGGING,
AEYQNIFTQVQVRGPADLGMTEDVNLANRSGVGPFSTLLGWFGNAQLGPIYLGSLGVLSLFSGLMWFFTIGIWFWYQAGWNPAVFLRDLFFFSLEPPAPEYGLSFA APLKEGGLWLIASFFMFVAVWSWWGRTYLRAQALGMGKHTAWAFLSAIWLWMVLGFIRPILMGSWSEAVPYGIFSHLDWTNNFSLVHGNLFYNPFHGLSIAFLYGS ALLFAMHGATILAVSRFGGERELEQIADRGTAAERAALFWRWTMGFNATMEGIHRWAIWMAVLVTLTGGIGILLSGTVVDNWYVWGQNHG
The first time you run an AlphaFold prediction the ChimeraX Google Colab panel will ask you to log in to your Google account to use their free servers. This is the same account for Google email, Googl Drive, and other Google services.
Google will warn that the ChimeraX AlphaFold code is not from Google. Press the Run Anyway button.
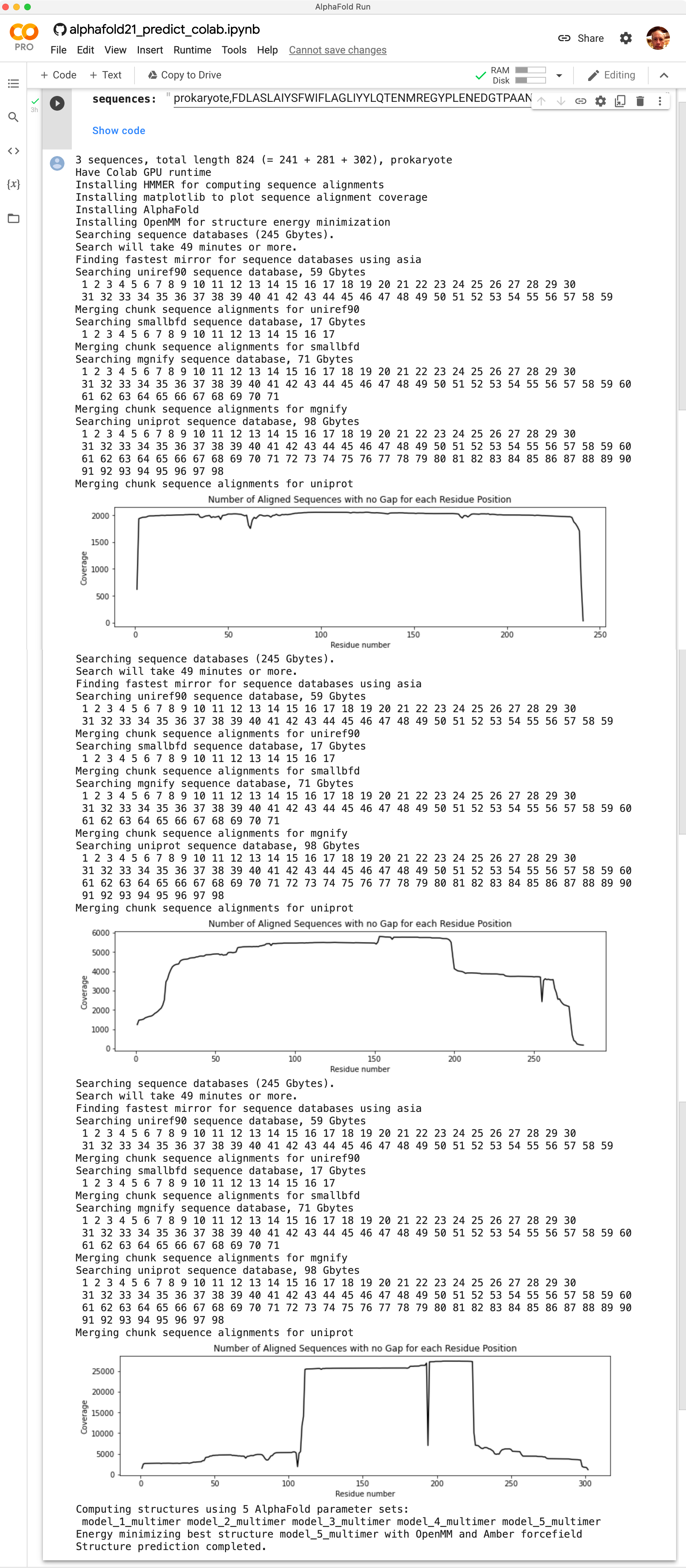
The AlphaFold Run panel is a web browser log of the output from Google Colab. It will show progress as each of the the 3 sequences is searched against databases of hundreds of millions of sequences and will show graphs of how deep the multiple sequence alignment is for each at each amino acid position. Sequence depths less than 100 can lead to poor predictions.
The best scoring of 5 predicted models will be opened in ChimeraX automatically when the calculation completes. All 5 models will be downloaded to directory
~/Downloads/ChimeraX/AlphaFold/prediction_1
After the AlphaFold prediction is loaded we can compare it to an experimental structure using the following commands.
open 6z1j
matchmaker #2 to #1
preset cylinder
To color the alphafold model by confidence (scores of 0-100 in the bfactor column of the PDB file) use the following command. Blue is high confidence, yellow, orange and red are low confidence. This example has a high confidence for almost all residues.
color bfactor #1 palette alphafold
To color the three proteins of the experimental structure with different colors use command
color #2 bypolymer
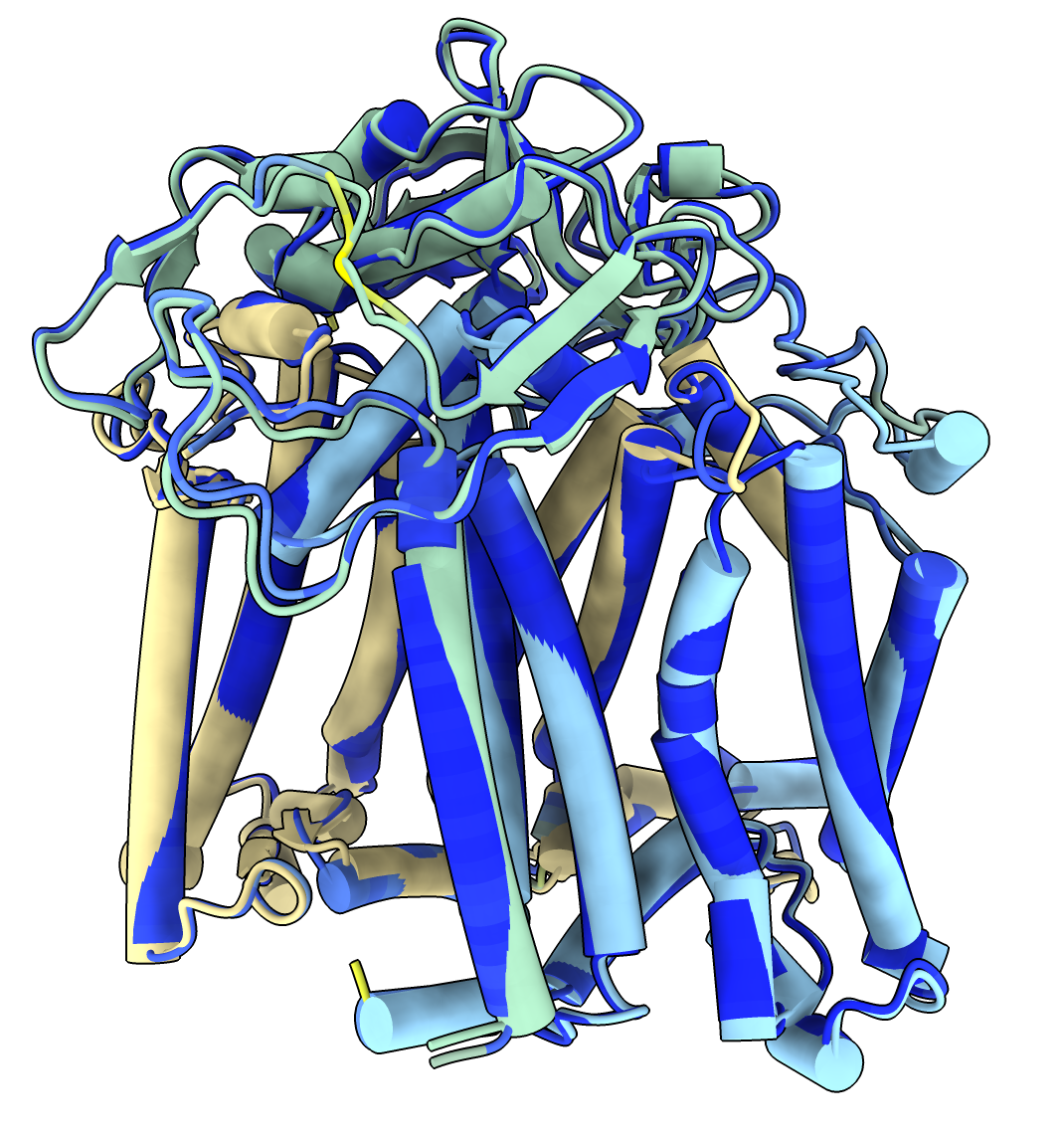 Image: Dark blue is alphafold prediction.
Image: Dark blue is alphafold prediction.
Light blue, green, yellow are the experimental structure. |