Work in progress November 21 - February 5, 2025
Tom Goddard
February 6, 2025
Group meeting
Topics
- Docking FDA drugs to viral targets with AlphaFold 3
- How to run AlphaFold 3 efficiently
- Alphafold 3 users
- Machine learning structure prediction ecosystem
- Virtual reality demos
- ChimeraX
Docking FDA drugs to viral targets with AlphaFold 3
abacavir sulfate, C1CC1NC2=C3C(=NC(=N2)N)N(C=N3)[C@@H]4C[C@@H](C=C4)CO.C1CC1NC2=C3C(=NC(=N2)N)N(C=N3)[C@@H]4C[C@@H](C=C4)CO.OS(=O)(=O)O
abametapir, CC1=CN=C(C=C1)C2=NC=C(C=C2)C
abemaciclib, CCN1CCN(CC1)CC2=CN=C(C=C2)NC3=NC=C(C(=N3)C4=CC5=C(C(=C4)F)N=C(N5C(C)C)C)F
abiraterone acetate, CC(=O)O[C@H]1CC[C@@]2([C@H]3CC[C@]4([C@H]([C@@H]3CC=C2C1)CC=C4C5=CN=CC=C5)C)C
abrocitinib, CCCS(=O)(=O)NC1CC(C1)N(C)C2=NC=NC3=C2C=CN3
... 1504 more
Drug docking results
- Runs took about 15 hours on Wynton for 1509 drugs against 300 amino acid viral protein targets
predicting 2 structures per drug + protein.
- Multiple sequence alignment for protein was computed once to speed up predictions 10-fold.
- A ChimeraX script sorted results by 3 Alphafold confidence metrics ipTM, pLDDT, PAE.
Monkey Pox E10R mRNA decapping enzyme example
Confidence scores
Name ipTM pLDDT PAE<3A/dist<4A/natom,nres,npair
estradiol 0.96 95 14 12 29
iptacopan_hydrochloride 0.95 95 28 18 46
efavirenz 0.95 93 20 15 48
doxycycline 0.95 94 32 14 66
citalopram_hydrobromide 0.95 89 19 13 34
...
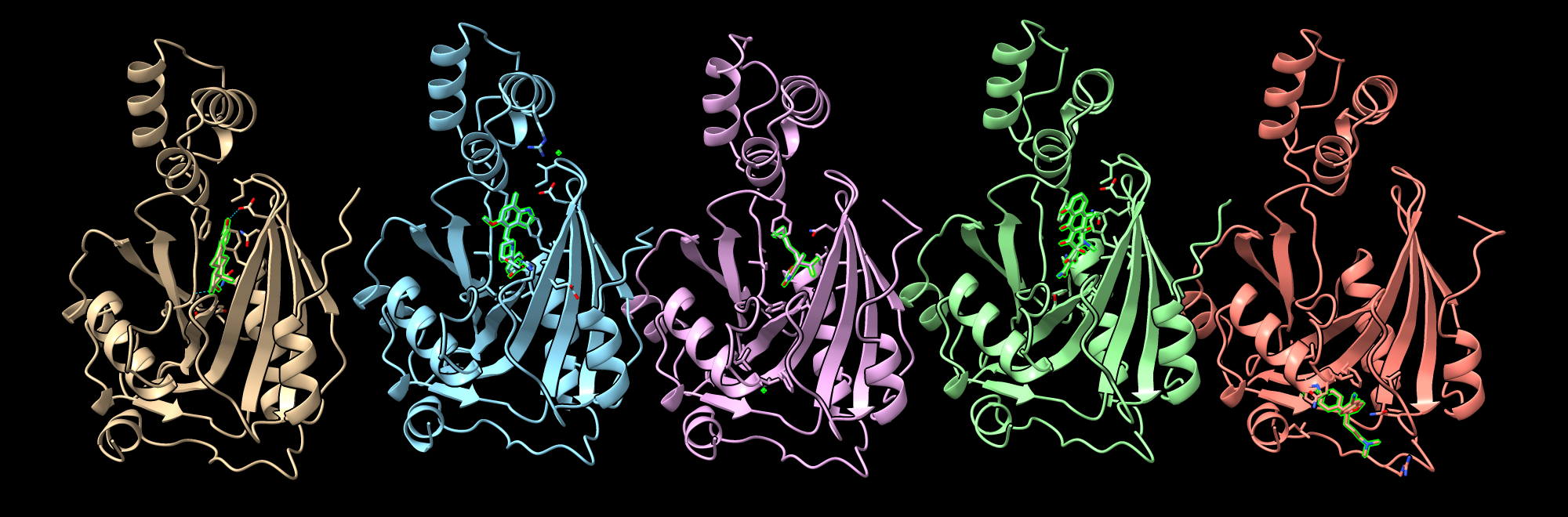
Poses of top 5 scoring ligands.
| 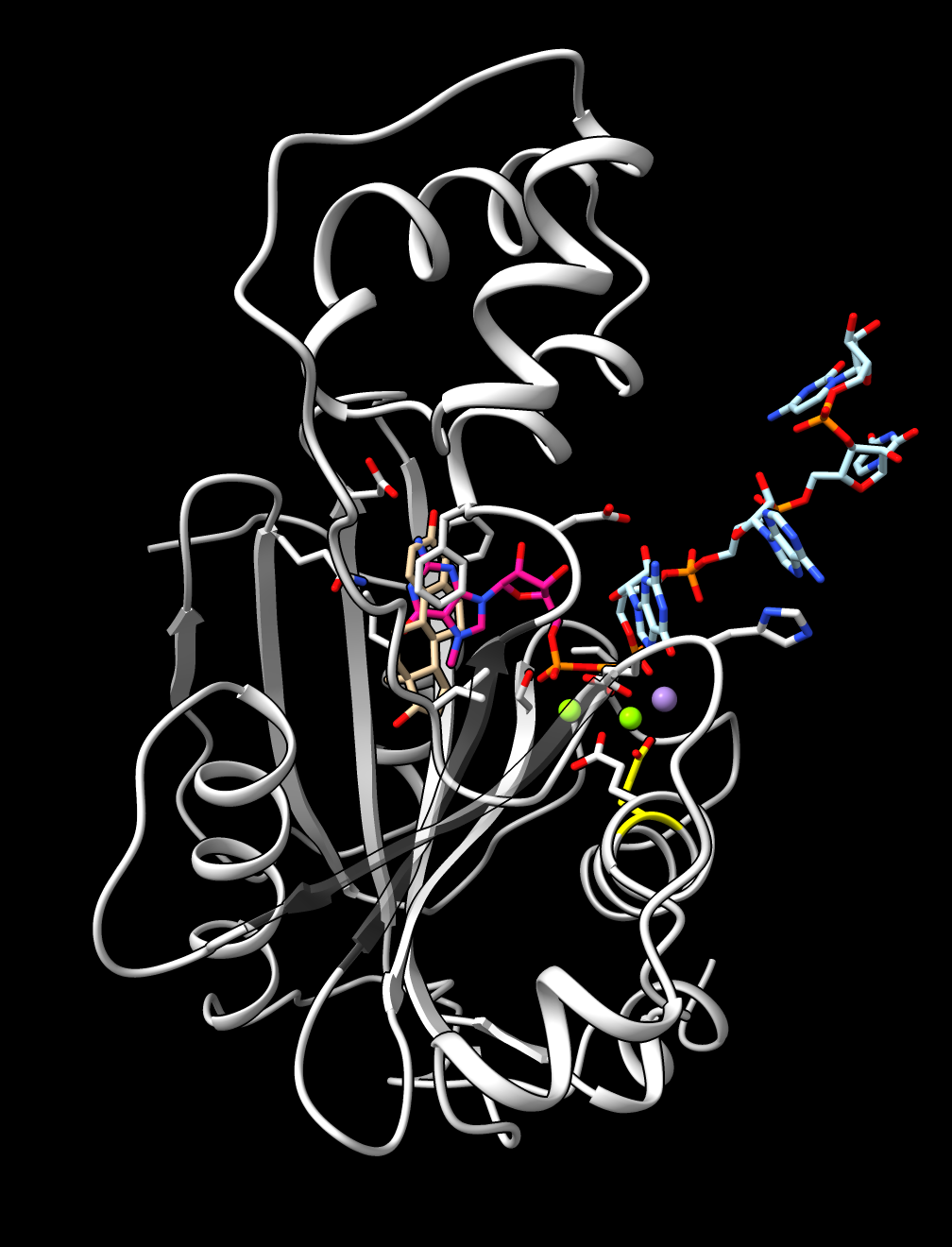
Native capped mRNA ligand
(cap pink, 4-mer mRNA blue)
with top drug (tan) superimposed.
|
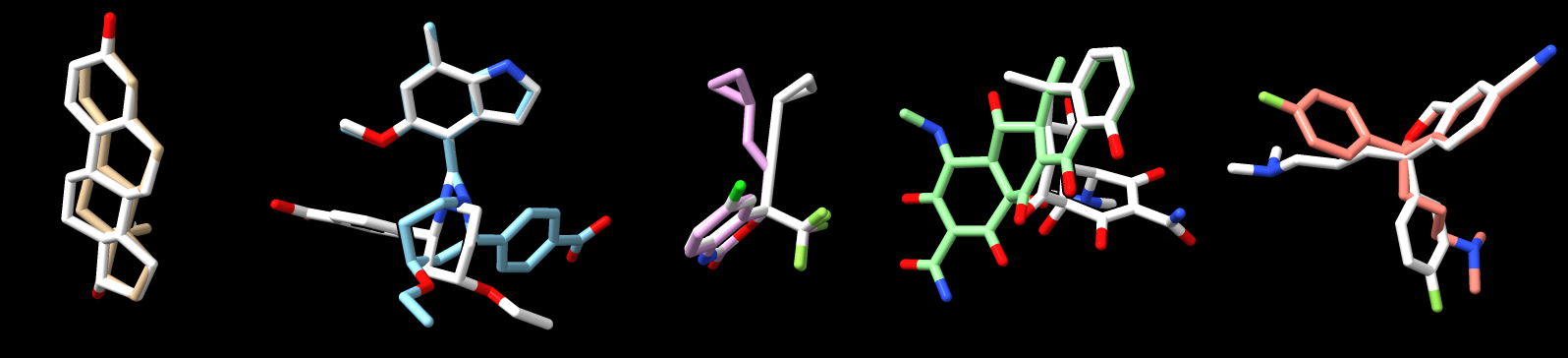
Two of the top 5 drugs had wrong chirality.
|
How to run AlphaFold 3 efficiently
- The AlphaFold 3 multiple sequence alignment (MSA) calculation takes 95% of the prediction time
for a 300 amino acid protein, 85% for 600 amino acid and 50% of the time for 3500 amino acids.
- The MSA calculation uses CPU only, no GPU, searching 300 Gbytes of protein sequence databases.
- Databases on an NVMe drive (minsky) is about 3x faster than the Wynton cluster BeeGFS for computing MSA
for a 600 amino acid sequence.
- RAM disk on wilkins was slower than NVMe drive because single-thread CPU speed limits maximum sequence
reading to 700 Mbytes/sec and wilkins CPU speed is half of minsky.
- I wrote a web page on
fast MSA calculation for AF3 describing the tests of different disks and CPUs.
- Most practical way to increase AlphaFold 3 prediction speed is to replace Jackhmmer sequence alignment
with faster mmseqs2 program.
| Disk | MSA time
seconds | Nvidia
GPU | Inference time
seconds | Machine | Notes
|
|---|
| NVMe | 293 | 4090 | 65 | minsky.cgl.ucsf.edu | CUDA 12.4, i9-13900K (8 performance cores, 16 efficiency cores)
|
| SSD sata 3 | 757 | 4090 | 73 | minsky |
|
| SSD sata 3 | 726 | 3090 | 104 | quillian.cgl.ucsf.edu | CUDA 12.6, i9-9900KF (8 cores)
|
| BeeGFS | 1049 | A40 | 124 | Wynton cluster qb3-atgpu30 | CUDA 12.4, AMD EPYC 7543P (32 cores)
|
Alphafold 3 users
- I made a singularity image for UCSF researchers to run AlphaFold 3 on Wynton and a web page explaining
how to run AlphaFold 3 on Wynton.
- Each user has to request their own set of AlphaFold 3 parameters (1 Gbyte) from Google.
Here are some researchers I have been talking with about how to make best use of locally installed AlphaFold 3.
- Adrian Pelin postdoc in Nevan Krogan lab - predicting pox-virus anti-viral tecovirimat against vaccinia virus protein.
- Andrii Kyrylchuk postdoc in John Irwin lab - running many predictions on Wynton.
- Liz Hong in Hiten Madhani lab - wants to run secreted fungal proteins against all human membrane proteins.
- Hiten Madhani - wants to predict a large methyltrasferase (> 2000 amino acids) against all cryptococcus proteins.
- Radhika Dalal graduate student in Tanja Kortemme lab - predict protein-dna interactions.
- Alex Pennachio in Yang lab at Gladstone - neuroscience and immunity, using AF3 on Wynton.
- David Fay, University of Wyoming - using Chai-1 alphafold 3 clone with ChimeraX PAE visualization, requested Chai-1 numpy PAE format to ChimeraX.
- Tristan Croll, Altos Labs - also using Chai-1 predictions and ChimeraX PAE plots.
- Willian Cortopassi, Gilead Sciences - Interest in Boltz an open source AlphaFold 3 clone from MIT. Google AlphaFold 3 is only for non-commercial use.
- Yaikhomba Mutum, University of Chicago - has predicted thousands of AlphaFold 2 dimers to study a mechano-sensation complex in hearing. Wants to use AF3.
- Xi Liu in John Gross lab, drug discovery.
- Billy Poon with Paul Adams, Phenix AlphaFold server running on LBL NERSC cluster.
Machine learning structure prediction ecosystem
Most of the useful machine learning structure prediction is being done by companies that restrict access
to their prediction software to try to make a profit. What can RBVI contribute?
- Developing ML structure prediction is currently too expensive for academic labs.
- Typical training uses 128 A100 80GB GPUs for a month for a single training. Development might involve 10 trainings.
- About $3 million in hardware and about $1 million operating training cost.
- Currently there seems to be only one competitive open-source ML structure prediction project, Boltz-1 from MIT.
Structure prediction projects
Here are a few of the well-known machine learning structure prediction projects and some of their use restrictions
and limitations.
Also see a
comparison of AlphaFold 3, Chai-1 and Boltz-1
from 30 November 2024.
- AlphaFold 3 code
and paper
- Unlikely to release advanced features such as ligand binding restraints.
- Slow MSA calculation based on 15 year old hmmer package.
- All users need to get their own AF3 parameters, 3 researchers asked me how to get them
after weeks of waiting with no response from Google.
- No training code, so no opportunity for others to improve AF3 network.
- No commercial use, so major user community excluded.
- AlphaFold 2 code
and paper
- Academic and commercial use allowed.
- Parameters freely available.
- Proteins only, no ligands or nucleic acids
- No training code, so improvements to network not feasible.
- Boltz-1 code
and paper
- Fully open source including training code and parameters, MIT license.
- Handles proteins, nucleic acids, ligands.
- Cannot handle large complexes (< 2500 aa with 80 GB GPU?).
- Uses various innovations beyond AF3.
- Can use experimental restraints.
- Chai-1 code
and paper
- Developed by hai Discovery
startup in San Francisco, $30 million funding for drug discovery.
- Handles proteins, nucleic acids, ligands, and binding restraints
- Cannot handle large complexes (< 2500 aa with 80 GB GPU?).
- Inference code available but not training code.
- Can predict with or without multiple sequence alignment.
- AlphaProteo paper
- Design of proteins that bind to other proteins.
- Code not available. Also no server.
- RosettaFold code
and paper
RosettaFold2NA code
and paper
RFDiffusion code
and paper
- Prediction of proteins, proteins and nucleic acids, and design of proteins that bind to other proteins.
- David Baker lab proof-of-principle software
- Lower quality results than commercial projects.
- Little code development after publication.
- ESM3 code
and paper
- Prediction and design of proteins, nucleic acids, ligands from Meta / Facebook, $142 million funding.
- Designed novel green fluorescent protein distant from known GFPs.
- Interesting design that handles sequence, structure and function keywords as input.
- Only small network is free. Published results use much larger network.
- Many other commercial projects... and smaller scale academic projects...

Nipah G protein with ligand zanamivir
PDB 8xps pink, AF3 green, Boltz blue, Chai tan
| 
AlphaFold 3 PAE
| 
Boltz-1 PAE
| 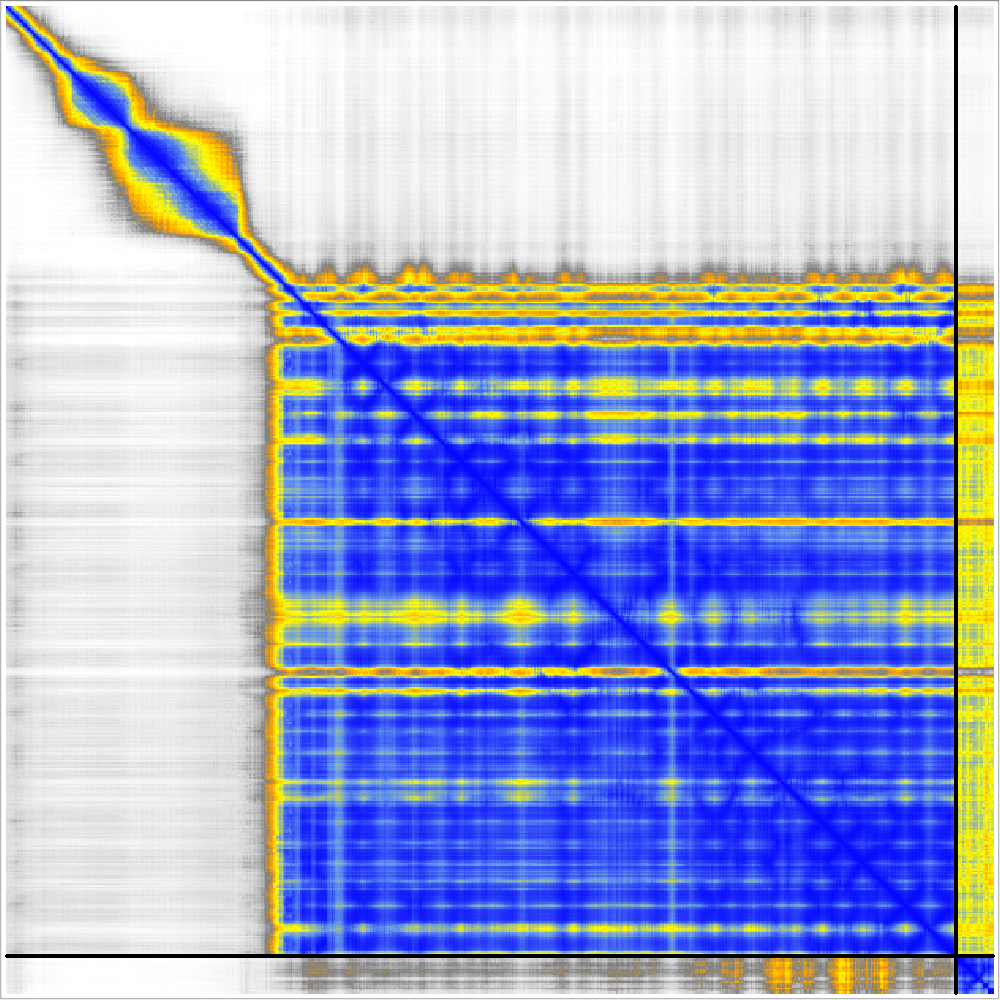
Chai-1 PAE
|
Virtual reality hardware
- Quest 3S standalone VR headset released October 15, 2024. Cheaper version of Quest 3 ($300 vs $500 for Quest 3), has cheaper lower-clarity Fresnel lenses and 10% lower resolution like Quest 2.
- Shiftall MeganeX superlight 8k
new PC VR headset, very light 179 grams (compare to 515 grams for Quest 3),
3500 x 3800 pixels per eye (similar to Apple Vision Pro), $1900, DisplayPort 1.4 cable only, tethered only, no wifi,
needs separate Vive base stations and wands, claimed shipping March 2025.
- Big Screen Beyond, light 200g, tethered only, high resolution 2500x2500 pixels per eye, $1600, requires lighthouse base stations.
- Acer SpatialLabs View Pro 27 inch auto-stereo (lenticular) 4k flat-panel display $3000, uses eye tracking and OpenXR interface. Krishnan at Biocryst offered to lend me his display. ChimeraX OpenXR may work with it.

Quest 3S
| 
MeganeX superlight 8k
| 
Big screen beyond
| 
SpacialLabs lenticular display
|
Virtual reality ChimeraX and LookSee users
Here are some people I have showed VR or helped with VR the past couple months.
- David Bhella, University of Glasgow - trying to use ISOLDE with ChimeraX PC VR, also using LookSee.
- Chris McClendon and Zoe Johnson, Pfizer - setting up standalone LookSee VR for researchers at Pfizer.
- Wilian Cortopassi, Gilead Sciences - he setup VR at Novartis but says Gilead is not interested.
- Ever O'Donnell and Willow Maestas-Coyote - vizvault demo of deep mutational scan coloring on ABC transporter.
- John Irwin and his son - VR demo in vizvault, 2 hours, including LookSee and ChimeraX. Wants ViewDock in VR.
- Andrii Kyrylchuk, postdoc in Irwin lab - vizvault demo viewing ligand binding.
- Jeff, Issac, Gushad and Joan from UCSF Machine Learning Masters program - vizvault demo.
- Student Cal and Doug Stryke - VR demo in vizvault looking at opioids.
- Le Minh, grad student in Dan Southworth cryoEM lab - says they recently got a Quest headset for molecules and cryoEM maps.
- Eric Tse, runs cryoEM facility in UCSF Sandler - borrowed two VR headsets to show LookSee at Sandler.
- Asmit Bhowmick, Lawrence Berkeley Labs - vizvault demo, plans to use LookSee for molecules.
- Bob Stroud - helped make default ChimeraX cross-eye stereo view have correct parameters (narrow field of view).

|
|
Other VR developments
- Met with NIAID team Meghan, Phil, David Liou, a few others on January 7 to discuss ideas for
an internal NIH proposal they were making for setting up an on-demand VR meeting server and possible
ChimeraX cloud computing. Don't know if they made a proposal.
- I tried to setup an RBVI Meta organization to get LookSee on the Meta app store using my
UCSF Id card (they requested driver's license or passport). They rejected it.
ChimeraX
- 1.9 release made, December 11.
- For 1.10 release in June 2025 considering replacing PyQt (commercially licensed) with PySide (free LGPL).















