
Tom Goddard
September 7, 2023
Group meeting

|
| IRAK4 dimer with inhibitor (PDB 8ATL). |
Answer:
Scooter has added the QScore reference.
Answer:
We improved the table 1 caption to now read: "ChimeraX extensions described in this paper. The developer who implemented the extension is listed and the first column cites the publication of the extension or original algorithm."
| 
| 
|
Answer:
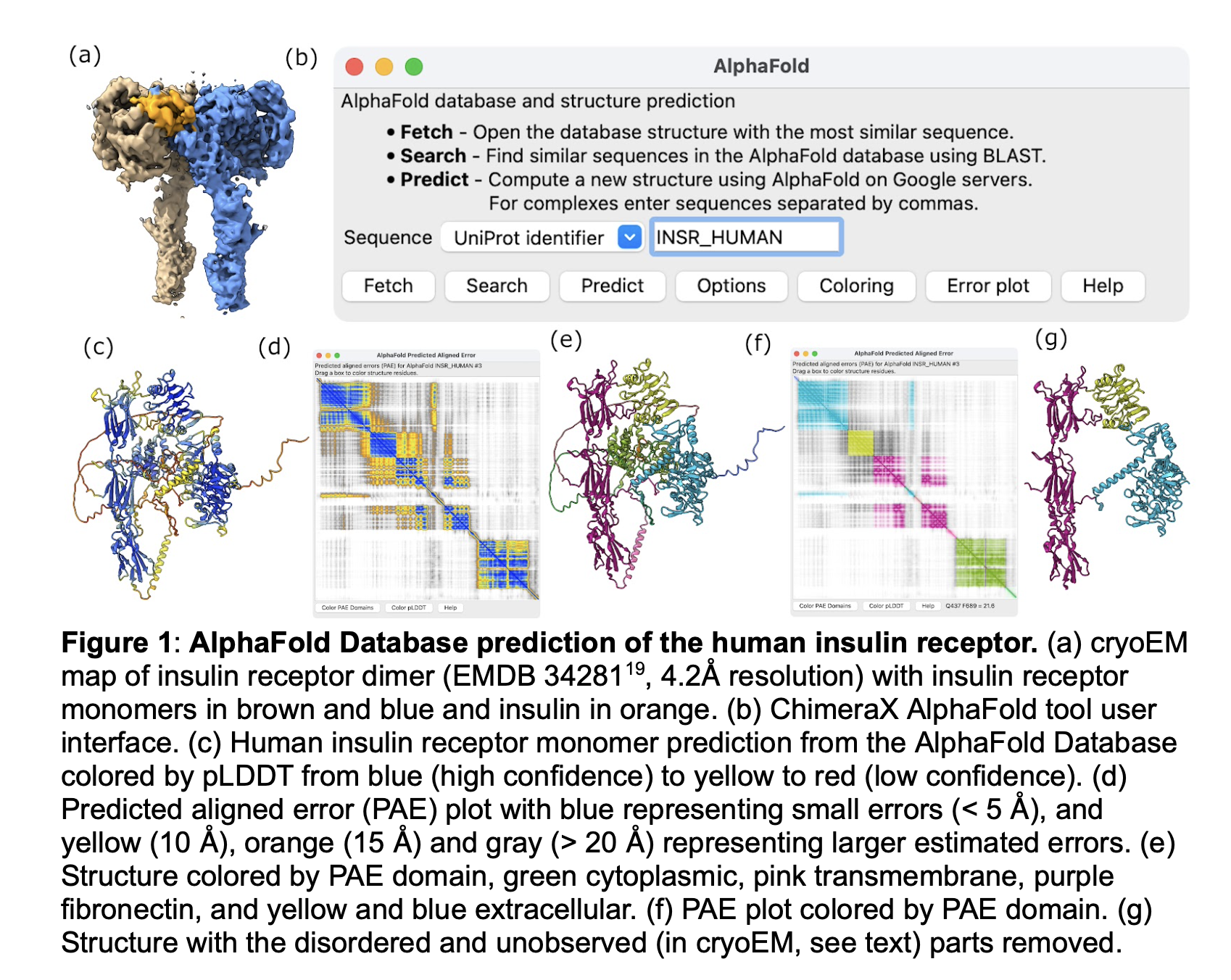
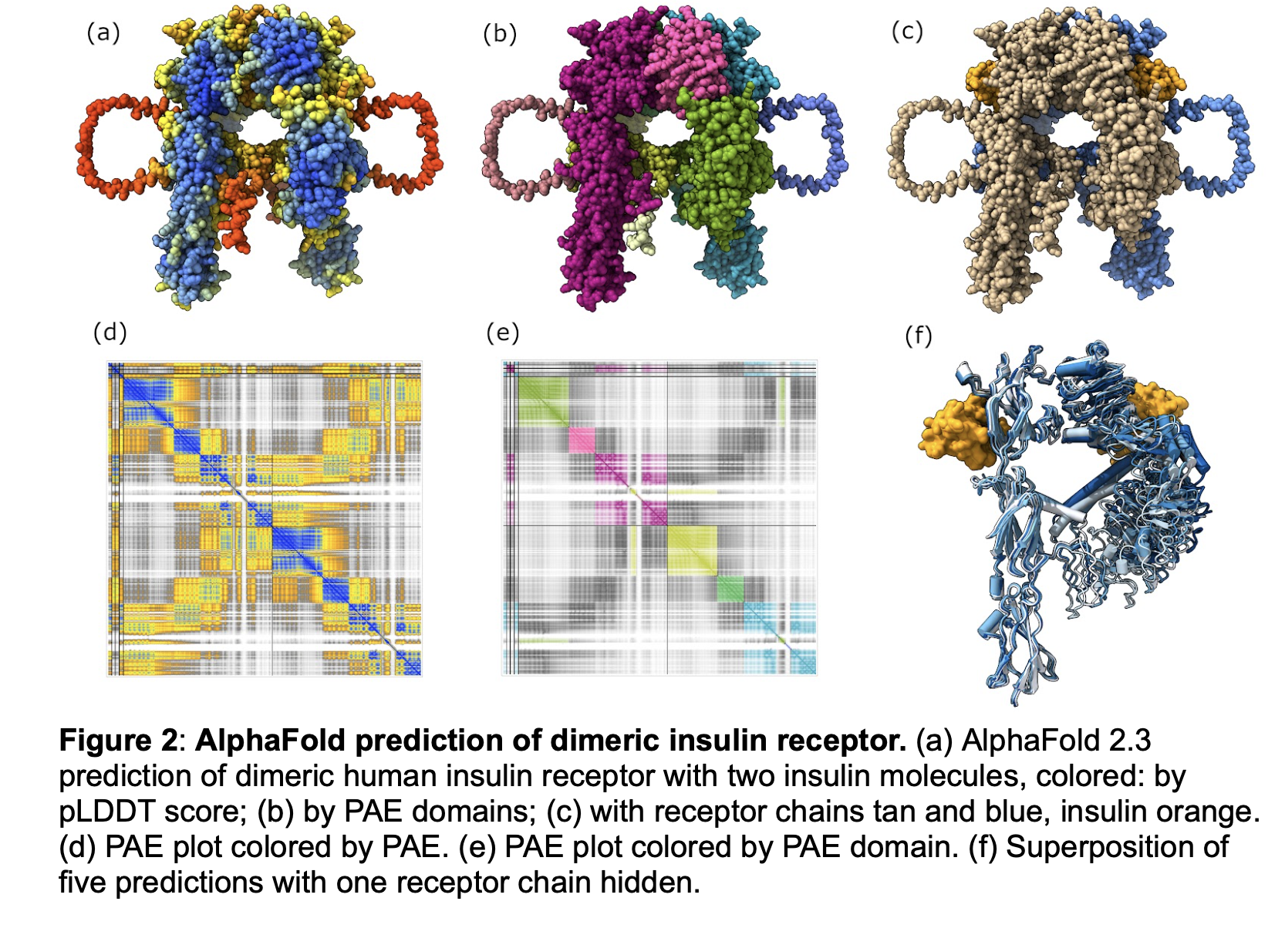
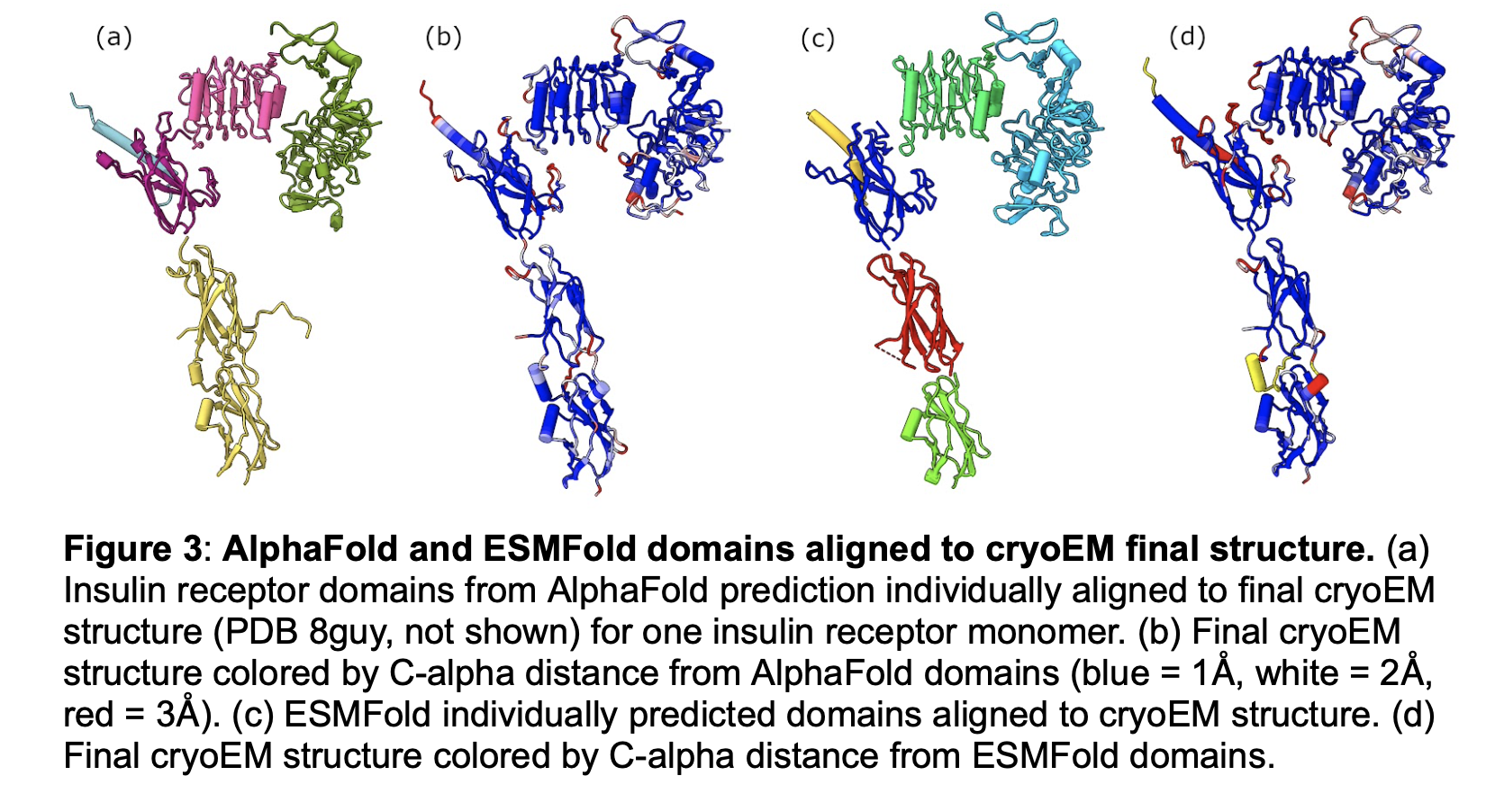
We will carlify the wording of figure captions making them more self-contained so they convey the purpose of the figure without having to read the main text of the paper. In figure 1 we can improve the c) description to say it does not match the cryoEM data, and in g) we can say it is the same as e but shows just the domains observed in cryoEM. In figure 2 we can explain f) better saying it shows the 5 ALphaFold predictions are very similar to one another, and refer to it as a "single monomer" of the dimer prediction. In figure 4 we can improve the bold title to say something like "Alphafold predicted insulin binding is at a secondary binding site". Also might say a-c are human to contrast with d mouse.
Answer:
We interpret this as a request that ChimeraX software be able to export the plots. It can currently export the PAE plots as a high-resolution PNG image and the ChimeraX colorkey command can make a suitable color key image that the user can compose into a figure. We agree being able to export a ready-made publication figure would be a nice feature.
Answer:
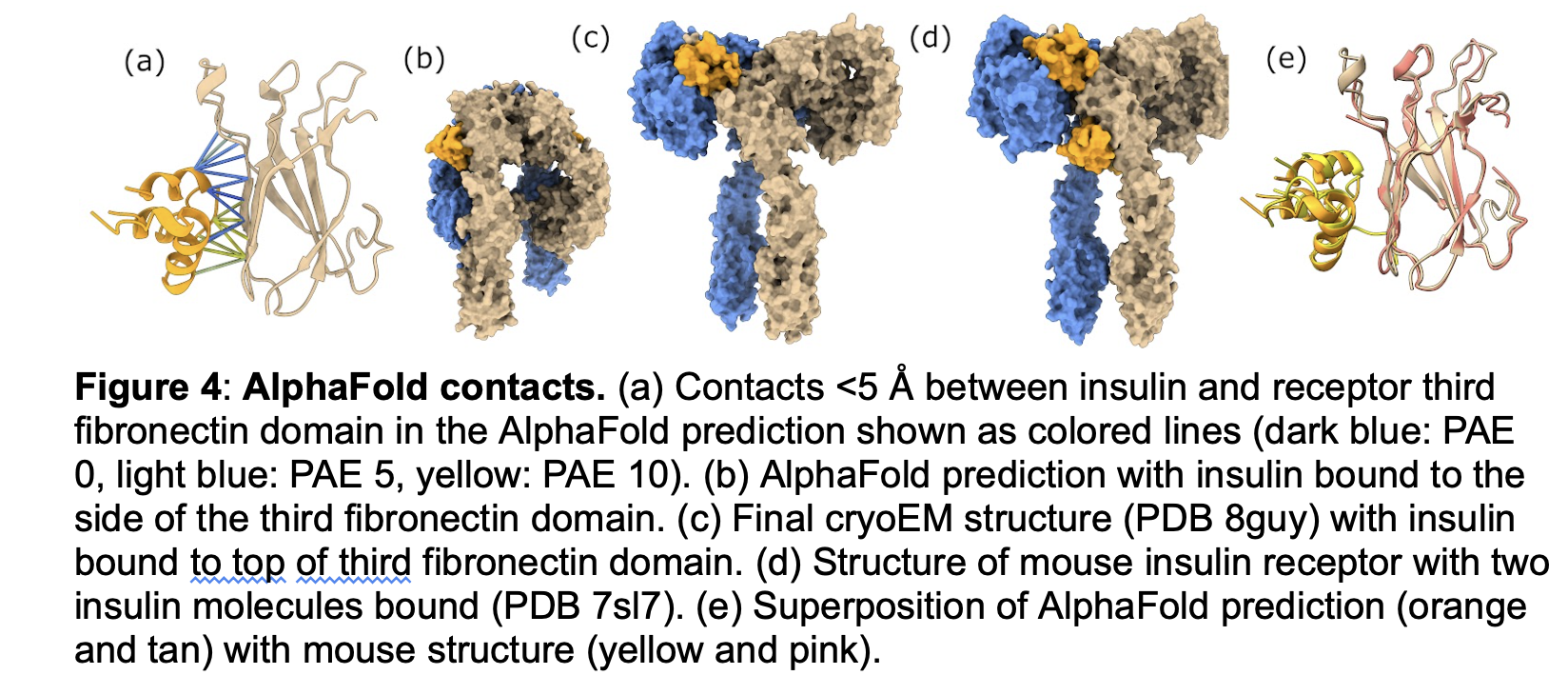
The "not shown" text is describing panel a), not panels b) and d). The b) and d) descriptions say they are the final cryoEM structure. It seems the reviewer missed that the "not shown" was in the a) description.
QScore section text:
"QScore approximates the side chain density by placing Gaussians at atom positions and correlates this predicted density to what is observed in the map. It only considers density closer to the assessed atoms than to any other atom. The Gaussian width is chosen to match very high-resolution maps (< 1.5A), so the value that is considered a good fit depends on the map resolution. This measure is the most widely used per-residue map fit score and is reported by the EM Databank for fit models. The ChimeraX extension can compute it for 1000 residues in 10 seconds and plot the scores. Clicking on a residue in the plot shows the corresponding residue and nearby map contour surface in the 3D graphics window."
Answer:
We will add text saying that QScore works on atoms other than amino acids. I checked that Tristan's QScore does work on nucleic acids, ions and ligands.
From the text:
"...aid understanding in vivo mechanisms such as the anticancer drug homoharringtonine."
Caption:
"Figure 7: Spatial arrangement of ribosomes seen in untreated cells and cells treated with anti-cancer drug homoharrington, with colors indicating the ribosome conformational state (Figure 4 of Xing et al. 202338). Ribosomes are more dispersed in drug-treated cells. "
Answer:
We will correct the caption to say homoharingtonine.
Answer:
This is already supported. ChimeraX extensions on the Toolshed often do log a message asking to cite their publication when used. Other extensions include in the user interface panel the citation. It is up to the extension developer to add this if they wish.
Answer:
We will use consistent hyphenation. Publications use both variants, although the big name cryo-EM labs seem to prefer the hyphen so we will use a hyphen.
Answer:
ChimeraX used the current AlphaFold version 2.3 for predicting the insulin dimer. While there are other prediction methods we believe we have used the latest state-of-the-art method, and of course the paper is limited to what is available in ChimeraX.
From the text:
"A new AlphaFold prediction (monomer or multimer) can be run on Google Colab servers by pasting the desired sequence(s) into the ChimeraX AlphaFold tool."
From the text:
"A maximum total sequence length of about 2000 residues can be predicted using a free Google Colab account. "
Answer:
We can add to the text when Google Colab is first mentioned that it requires a free account.