NIAID progress report for group meeting May 19 (last report) - July 14, 2022.
Tom Goddard
July 14, 2022
NIH 3D Pipeline
- Eric can describe progress.
- 3DPresets bundle installation issues.
DICOM
- Zach can describe progress.
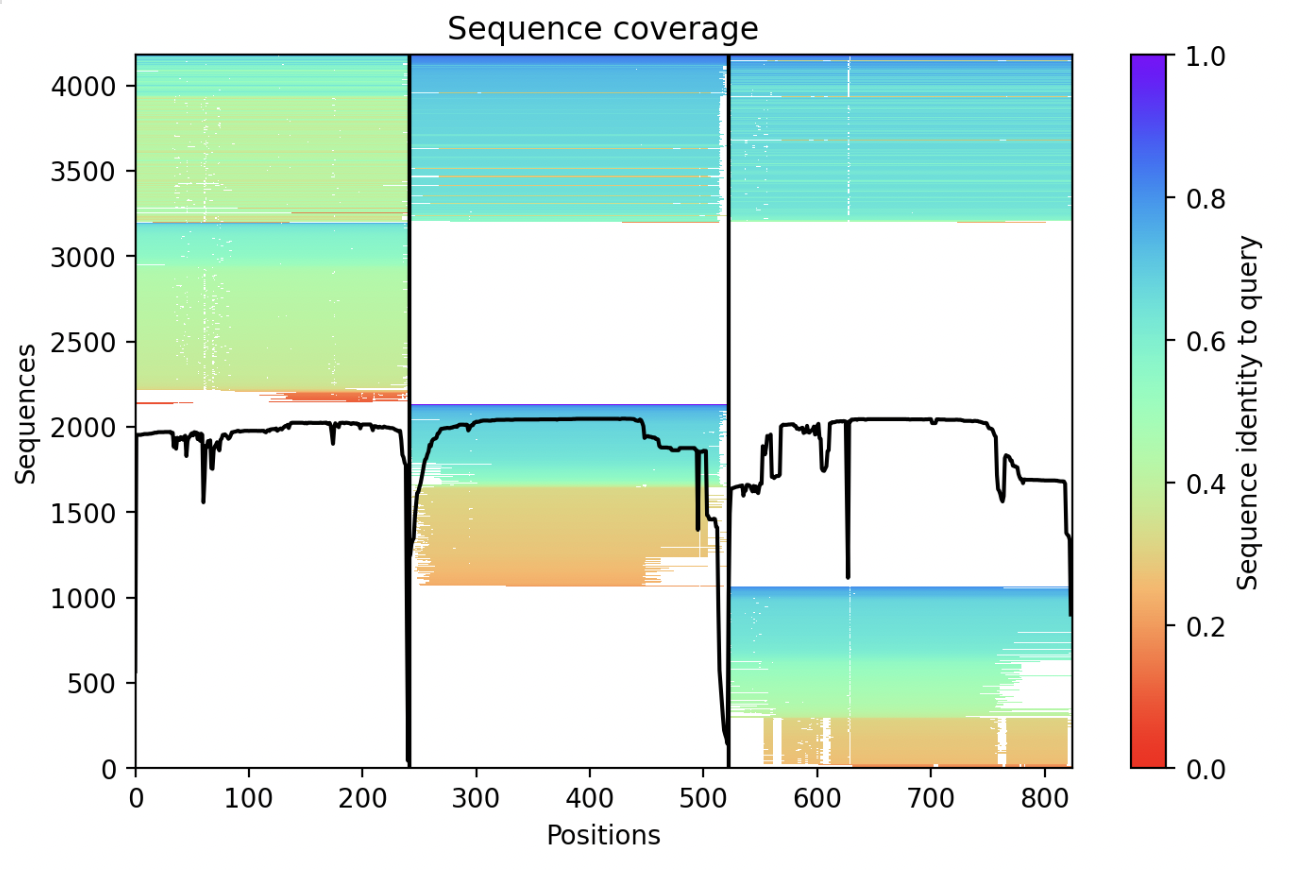
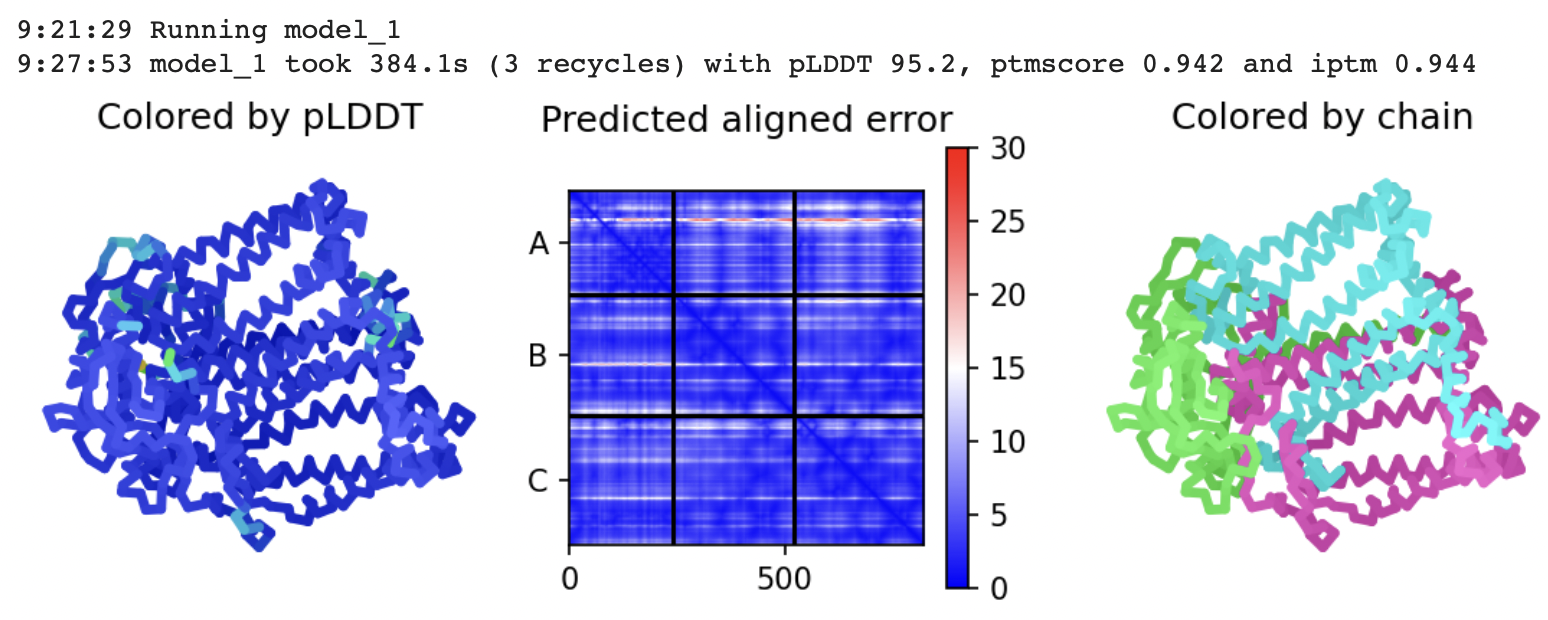
ColabFold: a 5-times faster AlphaFold
- Switching ChimeraX AlphaFold prediction to use ColabFold.
- Uses a separate server to make sequence alignment (48 cores, 768 Gbytes of memory) very fast,
hosted at Korean Bioinformation Center (KOBIC, similar to NCBI).
- ColabFold also runs the GPU structure calculation on Google Colab.
- Also optimizes expensive GPU compilation of neural nets.
- Also uses full size databases, and also can use PDB structure templates.
- Last 3 months, 12000 ChimeraX AlphaFold runs (140 per day), by 2700 unique IP addresses.
- Asked ColabFold developers Sergey Ovchinnikov at Harvard, and Martin Steinegger at Seoul National University if ChimeraX can use their server.
- Sergey and Martin are enthusiastic users of ChimeraX, responded immediately with a dozen emails.
- They are fine with ChimeraX using their sequence server and ColabFold.
- The ColabFold sequence server currently handles 12000 alignments per day from
the ColabFold web site.
- ColabFold uses a PyPi module on the Google Colab machine which also makes nice matplotlib plots that ChimeraX can show during the computation.
Native Mac M1 ChimeraX - Funded by CZI grant
- Setup nightly technology preview build on Mac Mini (euclid.cgl.ucsf.edu) including notarization.
- Made Mac universal distribution by combining ARM and Intel ChimeraX builds and tested.
- Still waiting for Phil Thomposon of Riverbank to fix PyQt6 commercial wheels. Said 2 days last week, now he says end of this week.
- Mac universal build (ticket #7222) dmg is 569 Mbytes (43% bigger) versus 398 Mbytes for Intel Mac dmg.
- Zach found LZMA compression can make ChimeraX dmg 30-40% smaller
(ticket #7274). Needs macOS 10.15.

