Tom Goddard
June 18, 2020
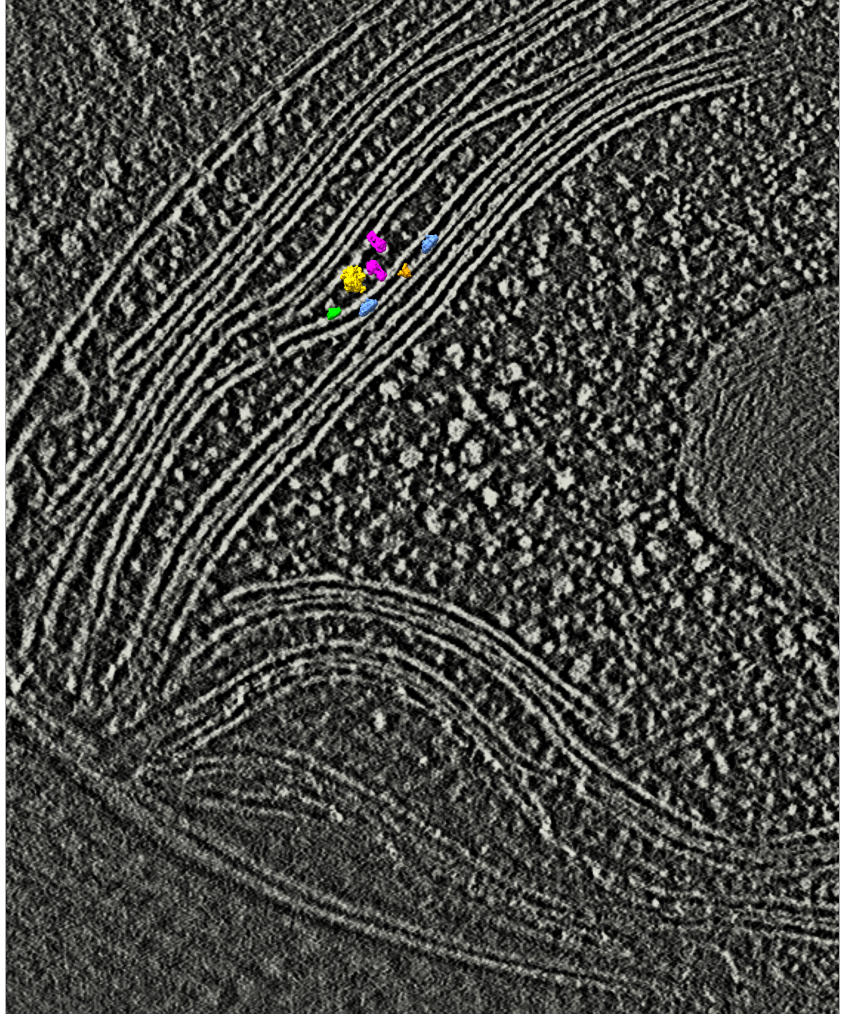
Try to place molecular complexes in EM tomography thylakoid membranes described in
Charting the native architecture of Chlamydomonas thylakoid membranes with single-molecule precision
Wojciech Wietrzynski, Miroslava Schaffer, Dimitry Tegunov, Sahradha Albert, Atsuko Kanazawa, Jurgen M Plitzko, Wolfgang Baumeister, Benjamin D Engel
Elife. 2020 Apr 16;9:e53740. doi: 10.7554/eLife.53740.
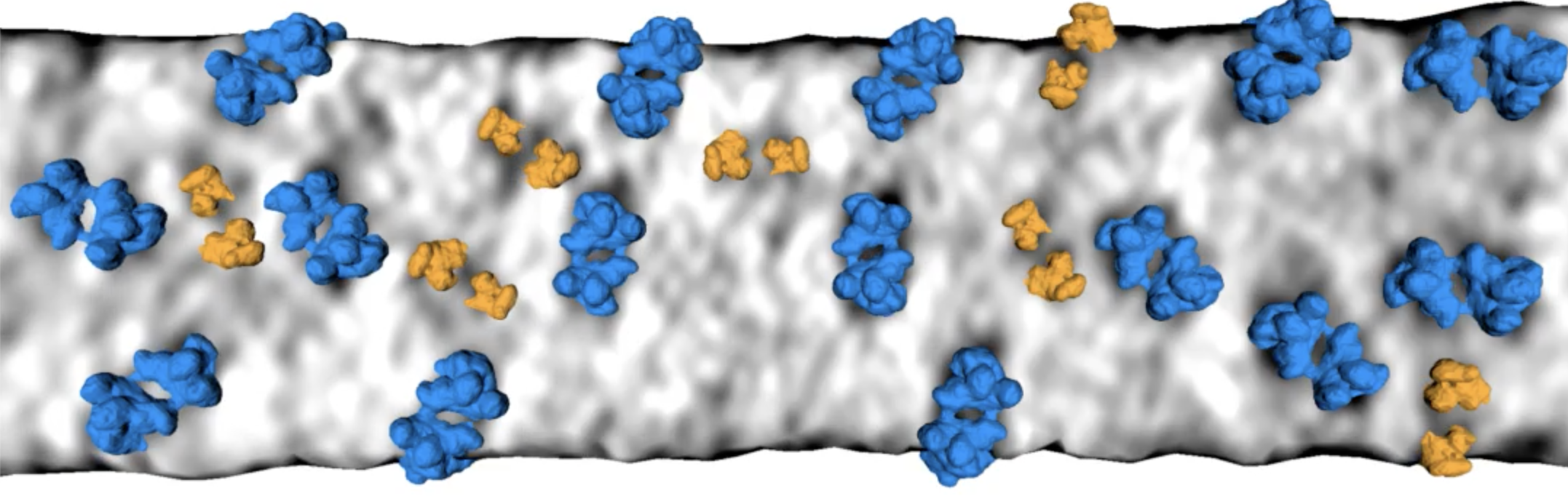
Use tomogram EMDB 10780. Invert so high map values correspond to high density (membranes and proteins are white instead of black, command "volume scale #1 factor -1").
Use molmap maps resolution 20 Angstroms for complexes:
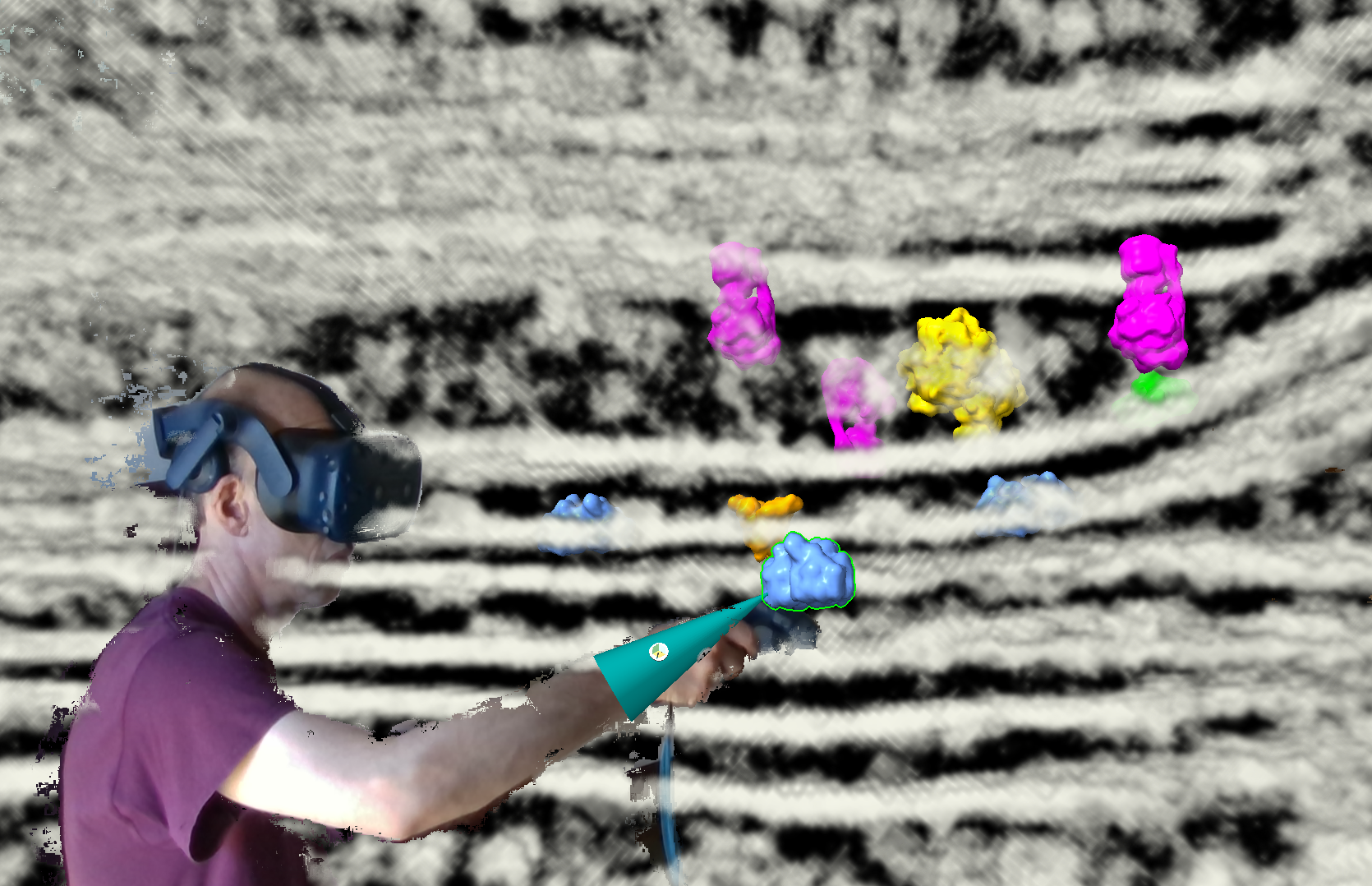
To easily duplicate and move molecules around I assigned several VR wand buttons.
The paper looks at graylevels on the membrane surfaces or a fixed distance out from the membrane surface. That is good because limits the complexity and noise. But it is also a bit misleading, suggesting the 3D density for the molecular complexes have nice shapes while in reality they are distorted, fragmented, difficult to recognize except by overall size and location relative to the membrane. The 2D membrane parallel images nicely capture the constraint of distance to the membrane and overall size and whether the density has two equal lobes. All this is harder in VR with 3D image display or surface display. While VR is workable, the 2D images give a clearer signal of whether a complex is present and which one it is.
The membrane parallel 2d images in VR may give the best of both worlds, good molecule recognition, and good appreciation of 3D arrangments on stacked membrane layers. The trick is getting the membrane surfaces.
Images from paper video 2: