
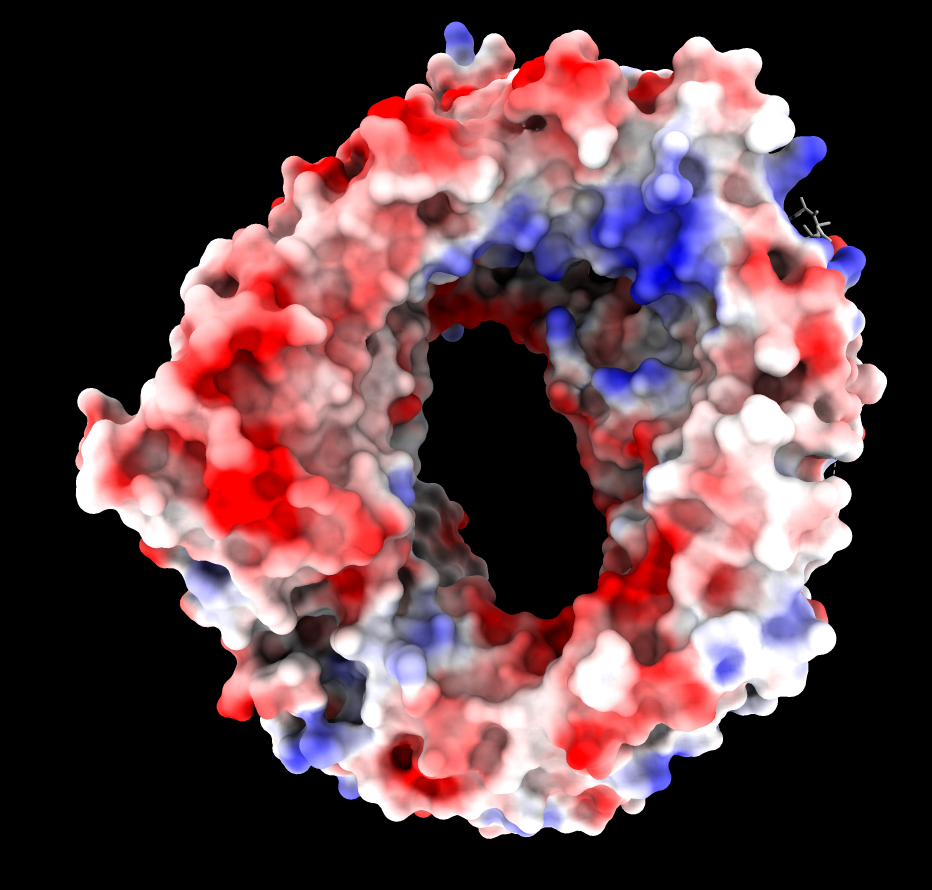
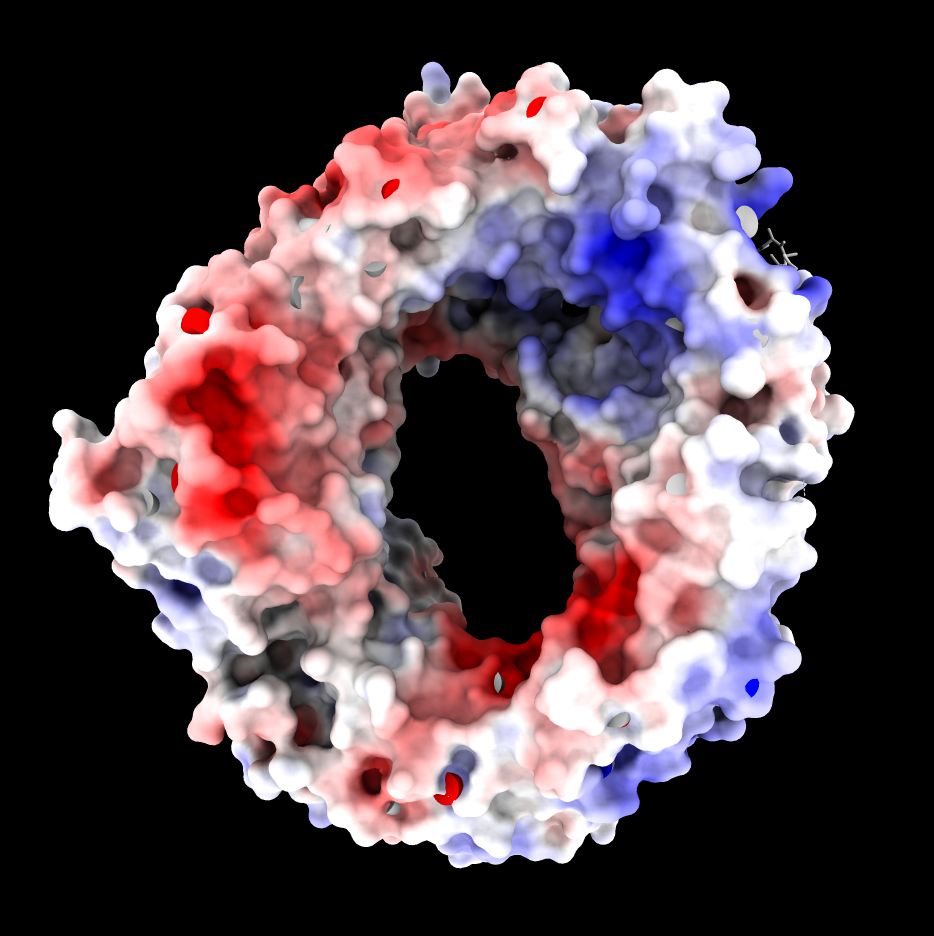
ASP and GLU negative charges (red).
ARG and LYS positive charges (blue).
Tom Goddard
April 20, 2021
Here is how to color a protein surface by electrostatic potential using ChimeraX or see the video.
Example will be nuclear export factor protein CRM1 which recognizes nuclear export signal sequences
on proteins that should be transported through nuclear pores out of the cell nucleus.
A positively charged patch on CRM1 is thought to be important in the release of cargo
once CRM1 gets out of the nucleus into the cytoplasm. The CRM1 biology is described in
Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo.
Another example of electrostatic coloring is given in the ChimeraX
protein-ligand binding site tutorial.
We look at electrostatics on PDB 6tvo which has CRM1 bound to Ran GTPase which enhances
affinity for NES proteins in the nucleus.
We also look at PDB 3gb8 which does not have Ran GTPase and
is in a conformation where binding affinity for NES tagged proteins is reduced, similar to
what is expected after release of cargo in the cytoplasm.
Fox AM, Ciziene D, McLaughlin SH, Stewart M.
J Biol Chem. 2011 Aug 19;286(33):29325-29335. doi: 10.1074/jbc.M111.245092. Epub 2011 Jun 27.
Models and Results

| 
| 
|
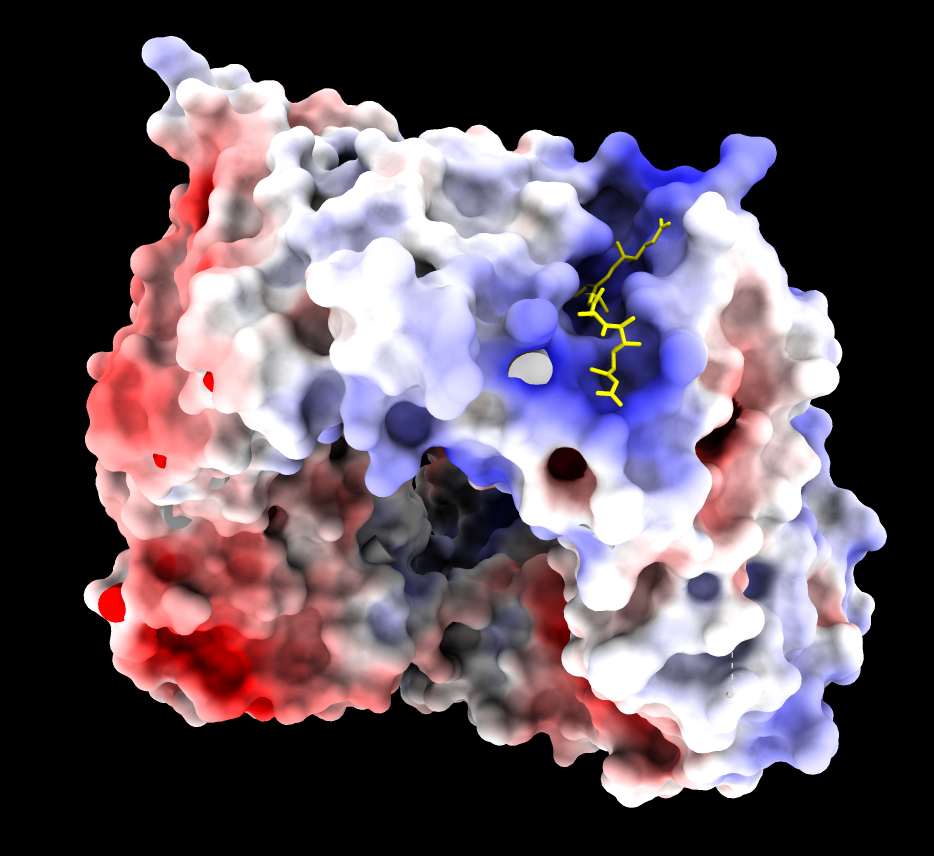
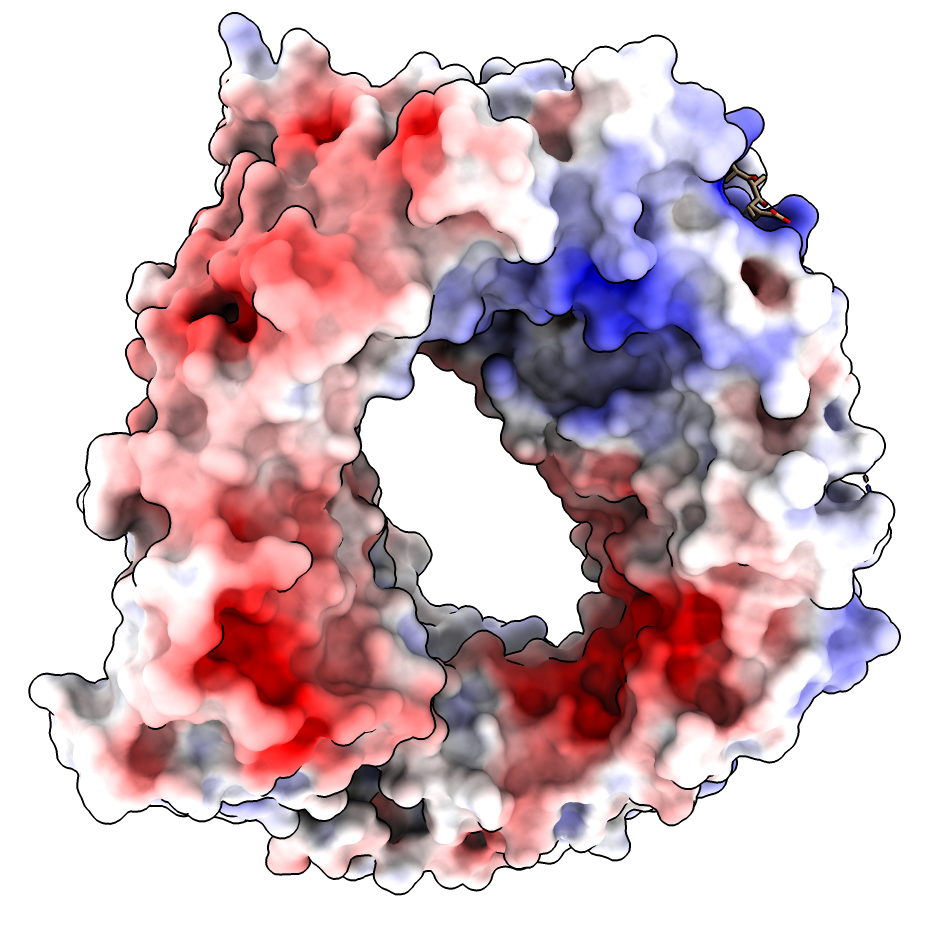
| CRM1 with Ran GTPase hidden, 6tvo. ASP and GLU negative charges (red). ARG and LYS positive charges (blue). | ChimeraX Coulombic coloring. | Adaptive Poisson Boltzman Solver (APBS) coloring. |
The blue (positive) patch in the upper right on the inside of the donut is thought to interact with the negatively charged C terminus of CRM1 stabilizing a state unfavorable for binding NES when Ran GTPase does not fill the donut hole.
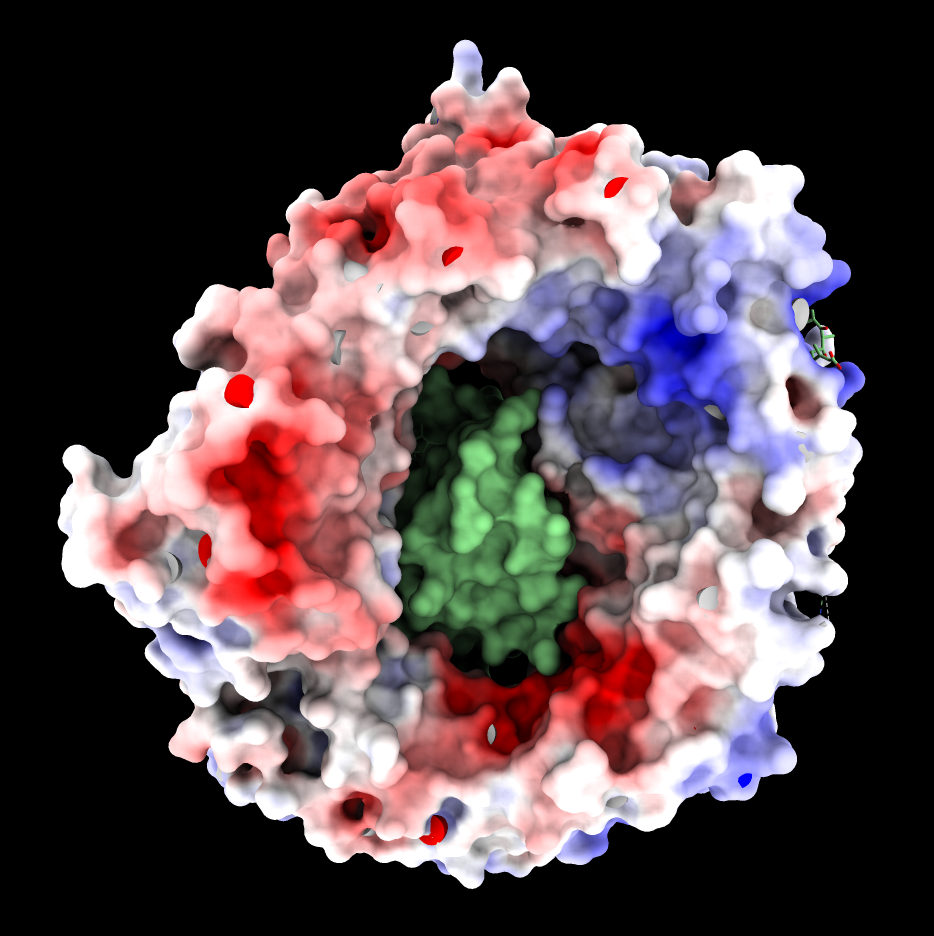
| 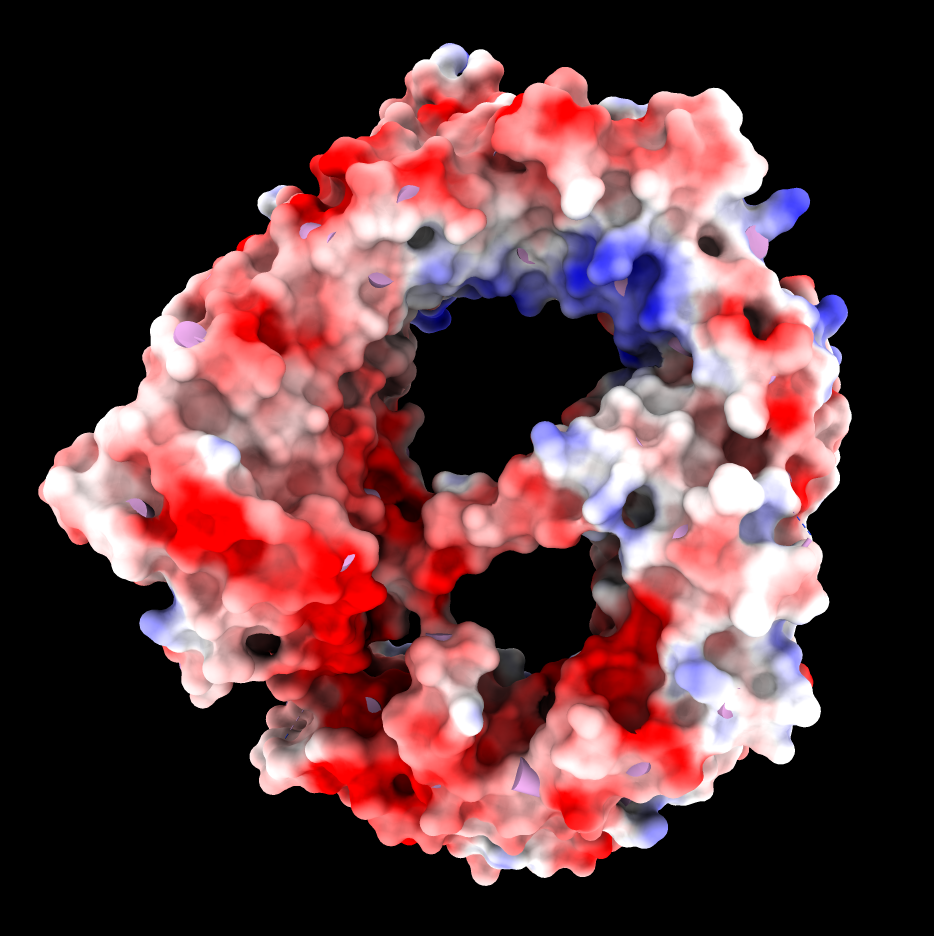
| 
|
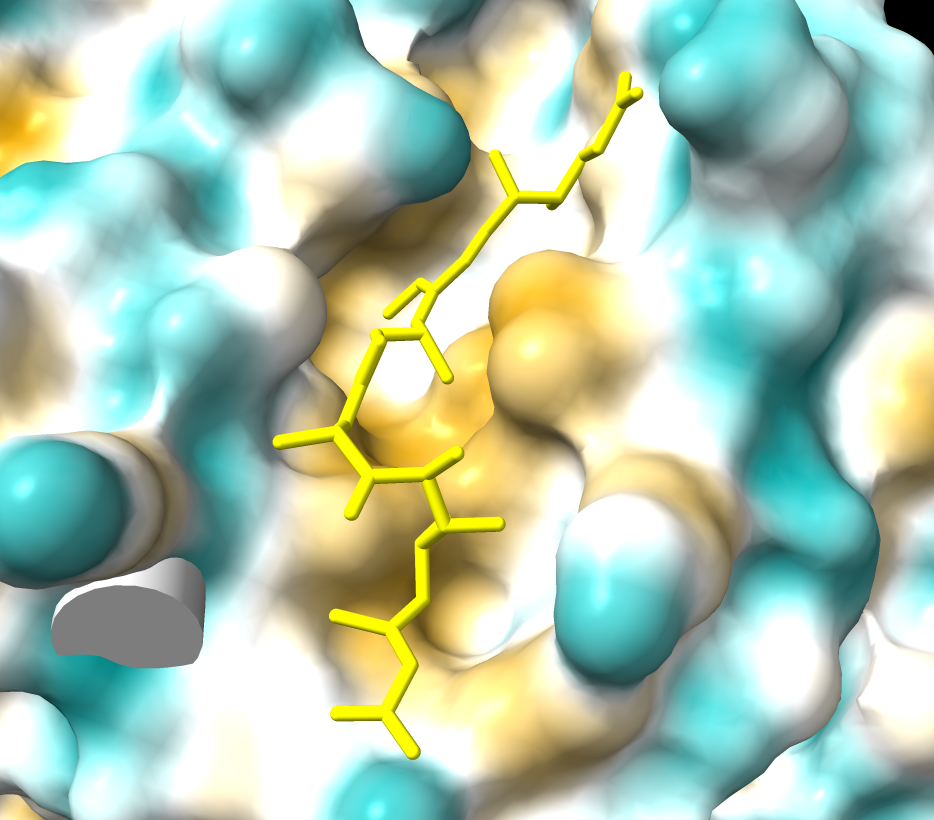
| CRM1 with Ran GTPase shown green, 6tvo. | CRM1 no RanGTP, C-terminal alpha helix crosses donut hole. PDB 3gb8 | Binding groove for NES is positively charged and has inhibitor Leptomycin B bound, and is hydrophobic (MLP coloring). PDB 6tvo. |
C-terminus 9 residues not resolved in structure 6tvo has 3 of 5 last residues negatively charged GLU/ASP (E/D) shown in ChimeraX sequence panel.

Ran PDB2PQR and APBS on web server using PDB 6tvo with RanGTP chain B deleted. This failed because PDB2PQR wrote a PDB file that APBS could not read. APBS does not adhere to the PDB format and requires the coordinates to be separated by white space and gives error that ATOM record 227 could not be parsed (ATOM 1542 CE LYS A 22 21.616 -49.085-100.520 1.00157.71 C). To work around this APBS bug I shifted the 6tvo structure by -50 in z in ChimeraX and saved a new PDB (move z -50 model #1 coord #1 ; save 6tvo_centered.pdb) to make the coordinates not exceed 100 Angstroms. Calculation took about 4 minutes producing a pot.dx file.
Here were the commands used to prepare the input PDB model for APBS.
open 6tvo
delete /B
move z 50 model #1 coord #1
save 6tvo_A_centered.pdb
The PDB2PQR web server added hydrogens and assigned charges producing PQR file s7822gmc58.pqr and also APBS input control file s7822gmc58.in.
The APBS web server then computed the electrostatic potential accounting for screening caused by polarizability of protein and solvent producing producing potential file s7822gmc58-pot.dx which is a 3d grid of electrostatic potential values in units kT/e.n
|
|
To color the CRM1 molecular surface by APBS computed electrostatic potential use the ChimeraX Surface Color tool available in March 2021 and newer ChimeraX versions. The steps are to open the atomic structure, compute the molecular surface, open the potential file, and color the surface according to potential values. All steps can be done using menus and user interface panels or with typed commands.
| Action | Notes |
|---|---|
| Home toolbar button Open | Open atomic model 6tvo_A_centered.pdb. |
| Molecule Display toolbar Surface show button | Shows solvent excluded molecular surface |
| Home toolbar button Open | Open APBS computed potential s7822gmc58-pot.dx. |
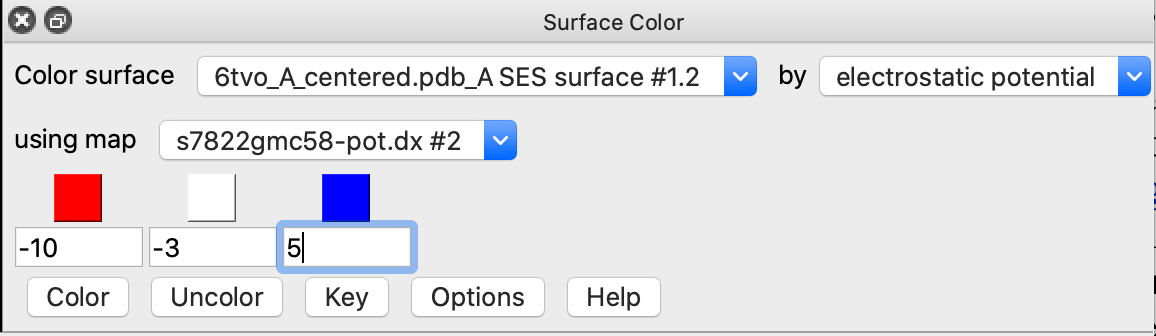
| Menu Tools / Volume Data / Surface Color | Color by electrostatic potential. Enter values below red, white, blue colors of -10, -3, 5. Press Color button. |
| Home toolbar button Soft lighting | This adds shadows which improves 3-dimensional appearance. |
| Model Panel, hide dx map. | Click off checkbutton next to s7822gmc58-pot.dx to hide potential contour surface. |
Equivalent ChimeraX commands
cd ~/Desktop/apbs-6tvo-files
open 6tvo_A_centered.pdb
surface
open s7822gmc58-pot.dx
color electrostatic #1 map #2 palette -10,red:-3,white:5,blue
hide #2 model
light soft
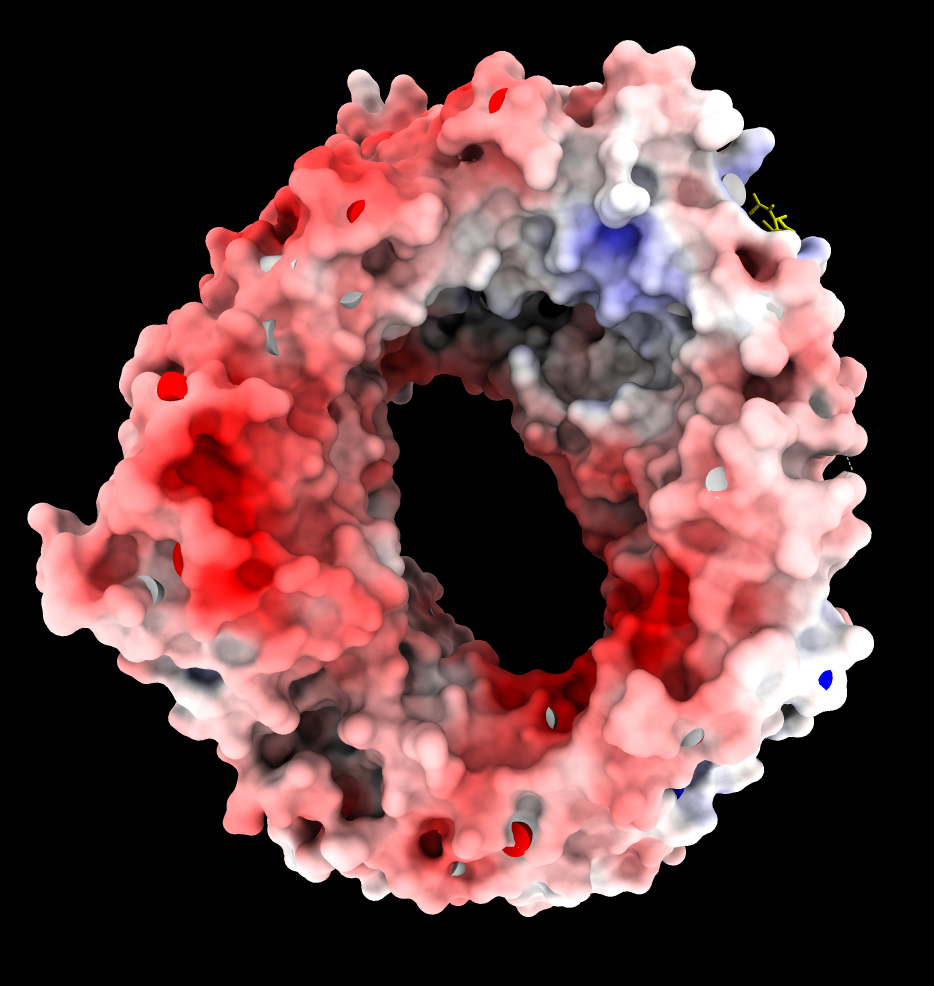
APBS produces an overall very red coloring using ChimeraX electrostatic coloring in Surface Color panel because there are 19 more negatively charged amino acids than positively charged. (select :GLU,ASP -> 115 residues, select :ARG,LYS -> 96 residues). These result in long range electrostatic interactions making most of the surface red. Mean ESP value on surfaces is reported as -3. Shifting red,white,blue colors to (-10,-3,5) instead of (-10,0,10) makes the coloring similar to Coulombic.
Why does Coulombic not have the same problem with mostly red coloring? It is using a hacky "distance dependent" dielectric potential V = q/(e*r) where e = C*r so the distance dependence is really 1/r*r which falls off much faster, so the positive regions are not masked by the long-range negative charges.
Is it reasonable to use the asymmetric ESP range for APBS coloring? I think so. Normally the 19 excess negative charged residues would be neutralized by ions at the surface so they would not cast long range effects. I guess explicit counter ions are generally not used in APBS calculations because they would strongly distort the potential near the ions.