

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
Molecular insights into capsular polysaccharide secretion. Kuklewicz J, Zimmer J. Nature. 2024 Apr 25;628(8009):901–909.
Bitter taste receptor activation by cholesterol and an intracellular tastant. Kim Y, Gumpper RH et al. Nature. 2024 Apr 18;628(8008):664–671.
Structure and assembly of a bacterial gasdermin pore. Johnson AG, Mayer ML et al. Nature. 2024 Apr 18;628(8008):657–663.
Structural basis of DNA crossover capture by Escherichia coli DNA gyrase. Vayssières M, Marechal N et al. Science. 2024 Apr 12;384(6692):227-232.
Molecular mechanism of actin filament elongation by formins. Oosterheert W, Boiero Sanders M et al. Science. 2024 Apr 12;384(6692):eadn9560.
More citations...News
January 22, 2024
ChimeraX 1.7.1 is available, with fixes for a few miscellaneous bugs that were identified after the 1.7 release.
December 19, 2023
The ChimeraX 1.7 production release is available! See the change log for what's new. Future Mac releases will require macOS 11 or higher.
November 6, 2023
The ChimeraX 1.7 release candidate is available – please try it and report any issues. See the change log for what's new.
Previous news...Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325, Chan Zuckerberg Initiative grant EOSS4-0000000439, and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Feature Highlight
|
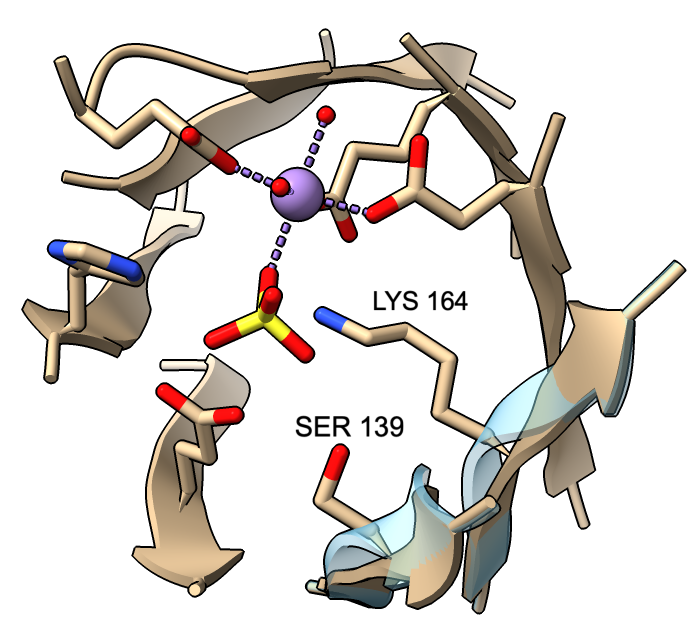
Cartoon ribbons are drawn to pass through the exact positions of peptide α-carbons in helix and coil, but by default, strands are smoothed to appear less ripply. This means that α-carbons in strands may not fall on the ribbon. A tether is drawn wherever a sidechain is displayed but would otherwise be detached from the ribbon. (Tether appearance can be adjusted with cartoon tether.)
However, β-strand ribbon smoothing can be tuned continuously between zero (off) and 1.0 (default, fully on), and this can be done for all residues or for specific residues only. The image compares ribbon positions for residues 139 and 164 in the active site of mandelate racemase (PDB 2mnr) with and without smoothing. The unsmoothed position (transparent blue) can be obtained with:
cartoon :139,164 smooth 0
This leaves ribbon smoothing at other residues unchanged. Values between 0 and 1 would give intermediate positions.
More features...Example Image
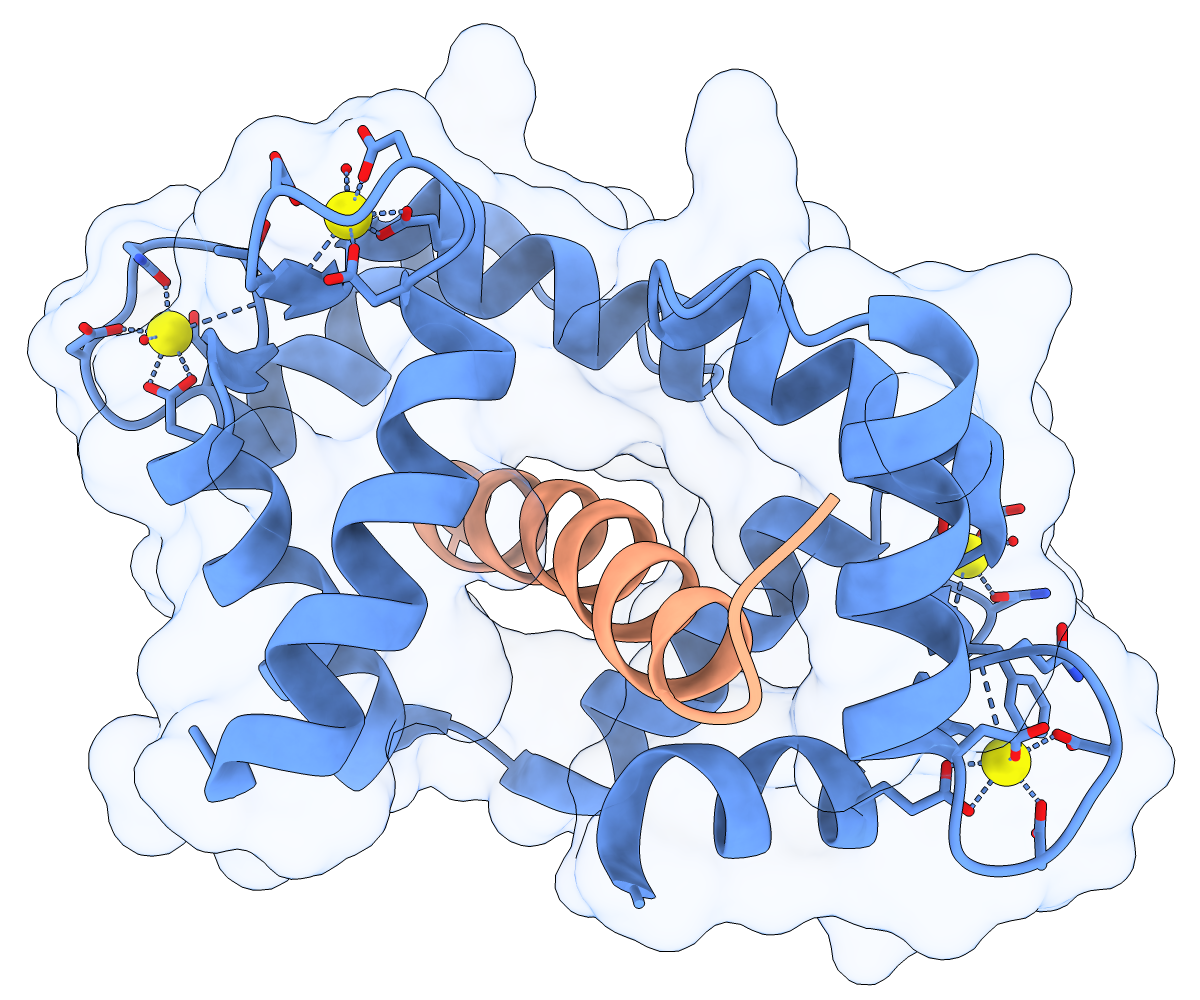
Calmodulin (CaM) acts as a calcium sensor. When its four Ca++ sites are fully occupied, it binds and modulates the activity of various downstream proteins, including CaM-dependent protein kinase I (CaMKI). Here, a complex between CaM and its target peptide from CaMKI (PDB 1mxe) is shown with cartoons, a transparent molecular surface, silhouette outlines, and light soft ambient occlusion. (If you prefer a less smudgy/rustic appearance, try using light gentle instead.) For image setup other than positioning, see the command file cam.cxc.
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.