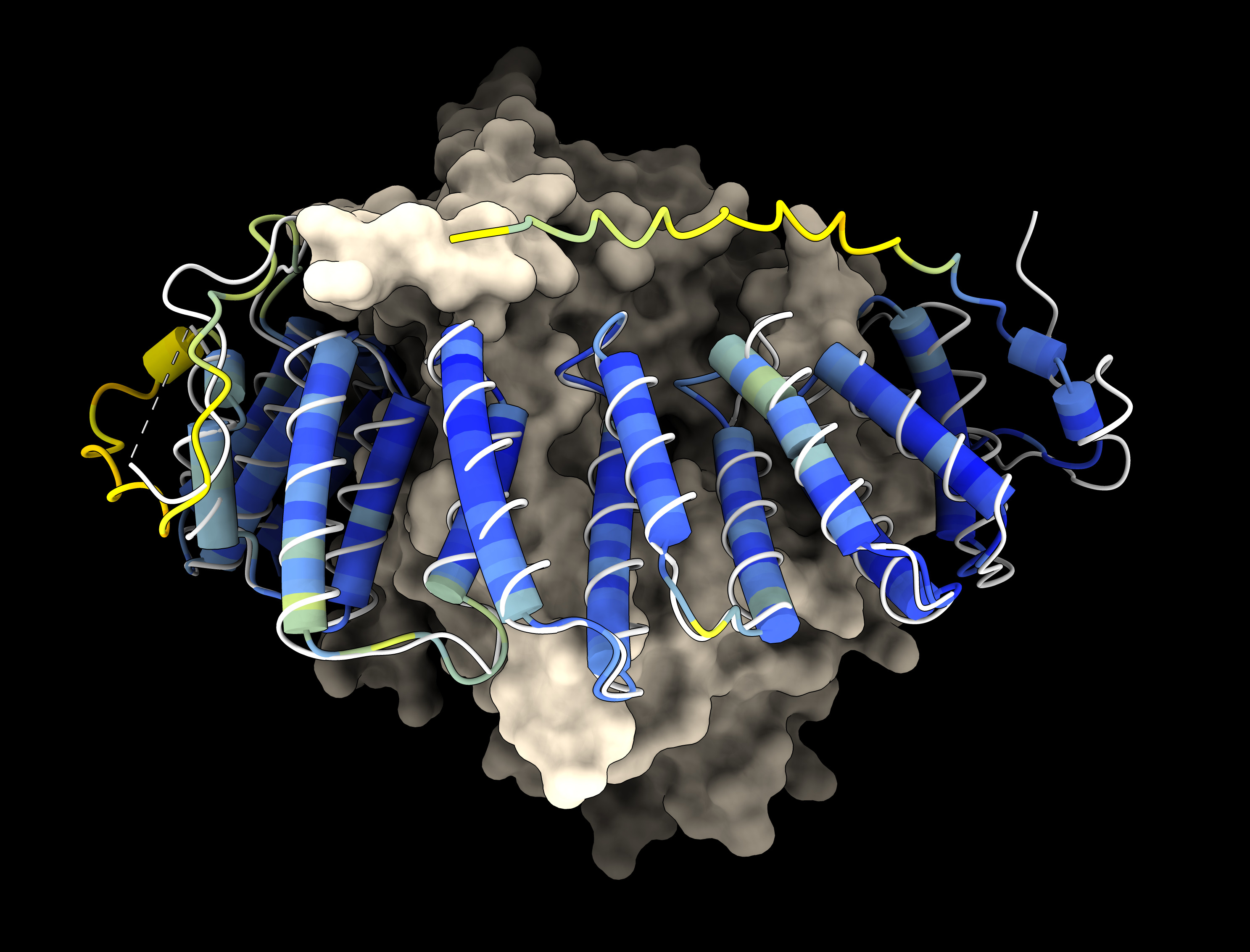
ESMFold predicted protein: farnesyltransferase subunit alpha colored by pLDDT confidence (blue, yellow) compared to experimental structure PDB 7t0a (white) with farnesyltransferase subunit beta shown as a surface.

ESMFold predicted protein: farnesyltransferase subunit alpha colored by pLDDT confidence (blue, yellow) compared to experimental structure PDB 7t0a (white) with farnesyltransferase subunit beta shown as a surface. |
Tom Goddard
November 9, 2022
You can predict a protein structure from a sequence using ESMFold (Evolutionary Scale Modeling) from ChimeraX (daily build from November 9, 2022 or newer, not in 1.5). ChimeraX uses the prediction server provided by the ESM Metagenomic Atlas described in this article
Evolutionary-scale prediction of atomic level protein structure with a language model
Zeming Lin, Halil Akin, Roshan Rao, Brian Hie, Zhongkai Zhu, Wenting Lu, Nikita Smetanin, Robert Verkuil, Ori Kabeli, Yaniv Shmueli, Allan dos Santos Costa, Maryam Fazel-Zarandi, Tom Sercu, Salvatore Candido, Alexander Rives
This prediction of farnesyltransferase subunit alpha takes about 20 seconds and uses the sequence from an experimental structure PDB 7t0a chain A.
open 7T0A
esmfold predict /A
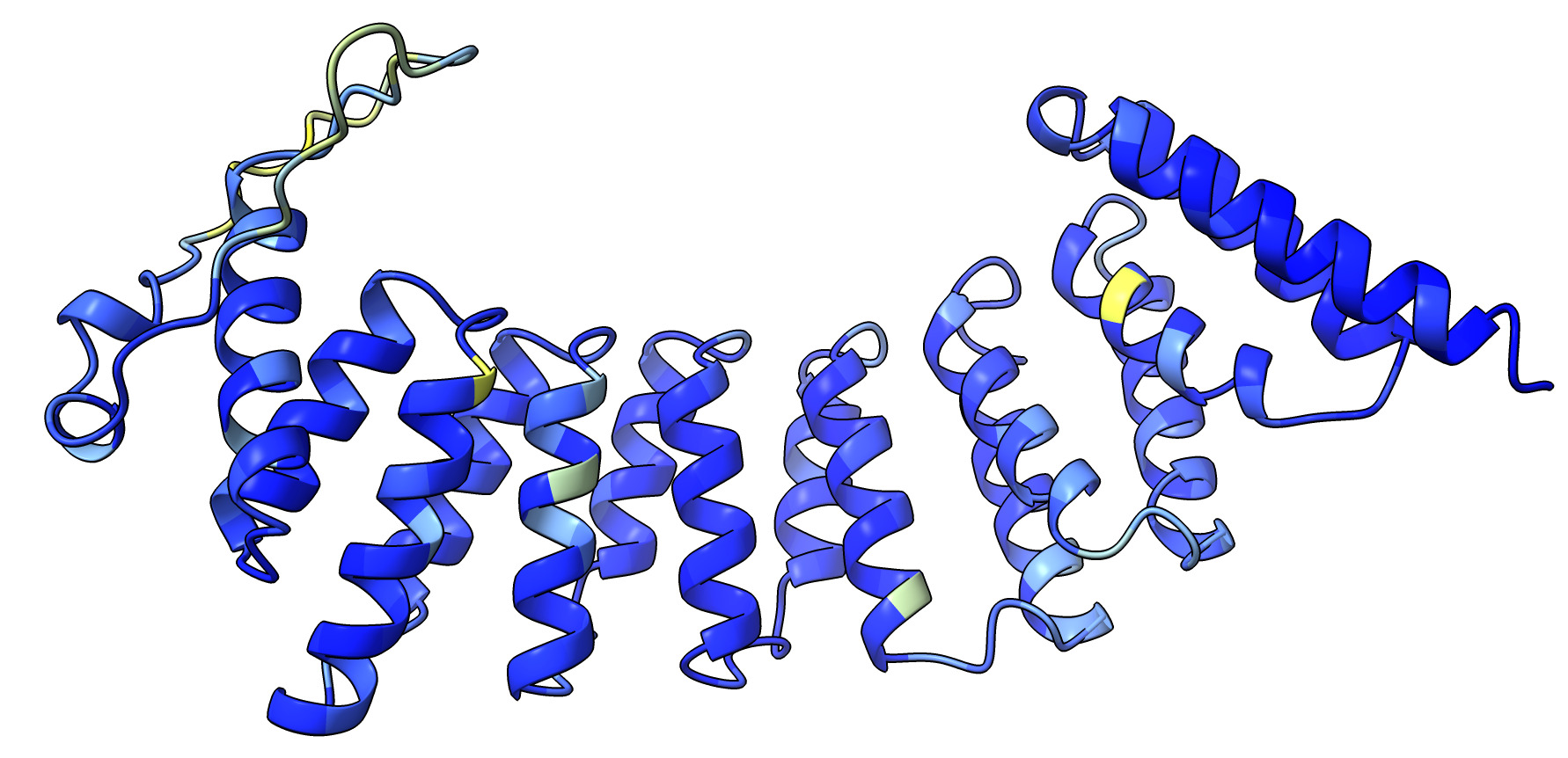
You can also make the prediction the sequence with no experimental structure.
esmfold predict MGSSHHHHHHSQDLMVTSTYIPMSQRRSWADVKPIMQDDGPNPVVPIMYSEEYKDAMDYFRAIAAKEEKSERALELTEIIVRMNPAHYTVWQYRFSLLTSLNKSLEDELRLMNEFAVQNLKSYQVWHHRLLLLDRISPQDPVSEIEYIHGSLLPDPKNYHTWAYLHWLYSHFSTLGRISEAQWGSELDWCNEMLRVDGRNNSAWGWRWYLRVSRPGAETSSRSLQDELIYILKSIHLIPHNVSAWNYLRGFLKHFSLPLVPILPAILPYTASKLNPDIETVEAFGFPMPSDPLPEDTPLPVPLALEYLADSFIEQNRVDDAAKVFEKLSSEYDQMRAGYWEFRRRECAE
| 
|
You can fetch a structure from the ESM Metagenomic Atlas in ChimeraX using the MGnify database identifier. To find identifiers a sequence search can be done at the atlas web site taking a few minutes. Predicted aligned error (PAE) can also be fetched.
esmfold fetch MGYP001094276757 pae true
esmfold predict /B residueRange 1,400
or you can automatically predict large sequences into chunks with some overlapesmfold predict #1 chunk 400 overlap 20