Boltz ligand binding predictions and Animation examples
Tom Goddard
May 23 - August 14, 2025
Group meeting
Topics
- Boltz structure prediction evolution in ChimeraX
- Batch ligand predictions for drug discovery
- Boltz ligand prediction demo
- When are ligand predictions accurate?
- Prediction consistency with different random seeds
- Future ChimeraX Boltz improvements
- Animation examples
- Animation tips
Boltz structure prediction evolution in ChimeraX
- Boltz is a structure prediction program derived from AlphaFold 3 with similar accuracy to AF3 predicting assemblies of proteins, DNA, RNA, ligands, ions, solvent.
- ChimeraX installs and runs it on local Mac, Windows or Linux computer.
- ChimeraX Boltz tool web page, and User Guide documentation.
- July 11, 2025: Boltz 2.1 with ligand affinity used by ChimeraX in daily builds, how-to-use video posted on YouTube and BlueSky and X.
- June 26, 2025: Boltz 1.0 used by ChimeraX 1.10 release with 16-bit floating point precision I added for handling larger structures, and steering potentials to improve geometry.
- April 18, 2025: Boltz 0.4.1 added to ChimeraX. How-to-use video on Youtube and BlueSky and X.
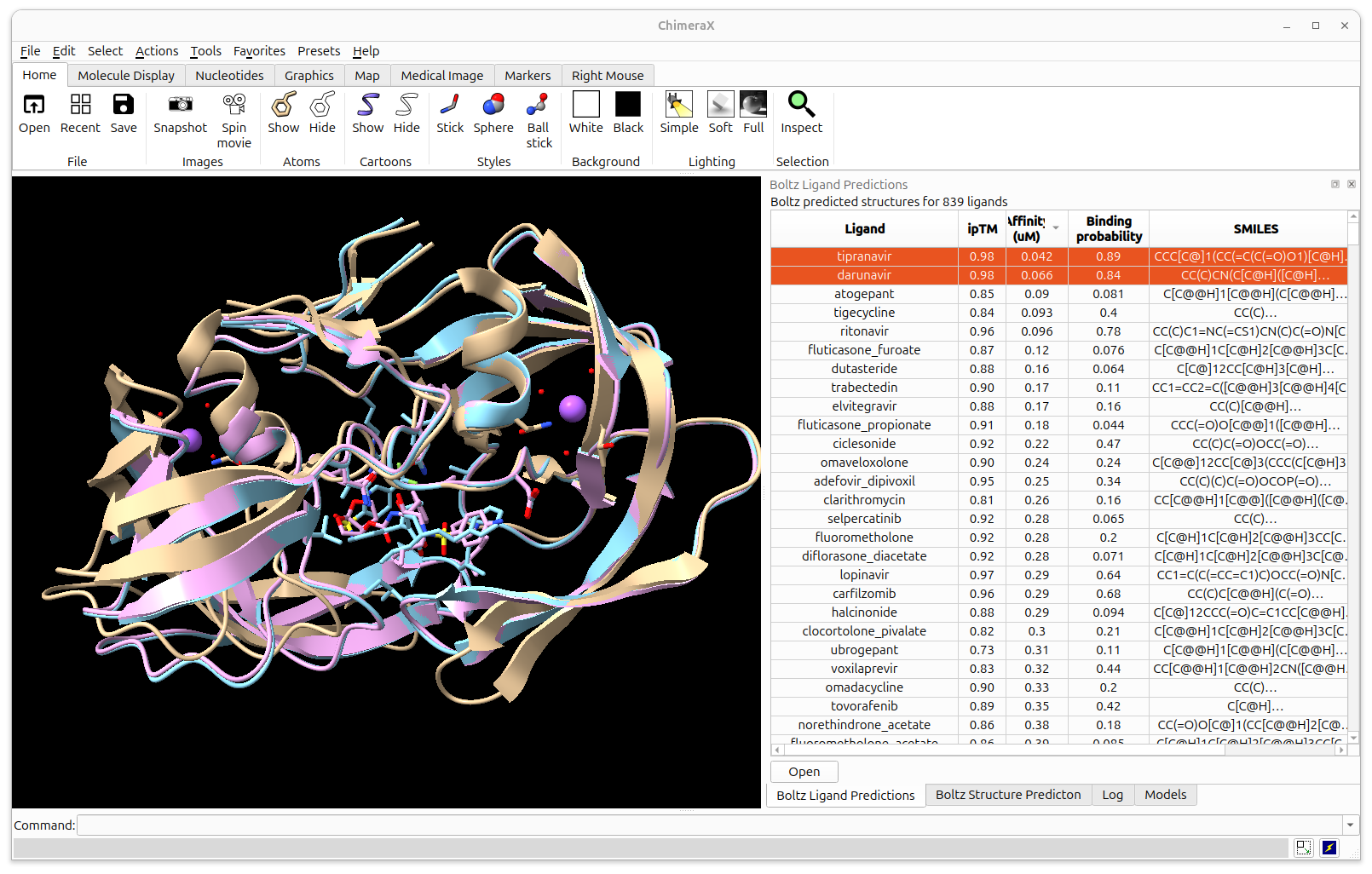
Next ChimeraX Boltz improvement:
batch ligand predictions for drug discovery
- Demonstrate how to setup and run batch prediction.
- ChimeraX can setup and run batch ligand predictions against protein targets using Boltz 2.2.
- 839 FDA compounds run against HIV protease dimer took 3 hours on Nvidia 4090 (Linux), 24 hours on Mac M2 Ultra.
- Plan to release in ChimeraX daily build in coming week and post example video.
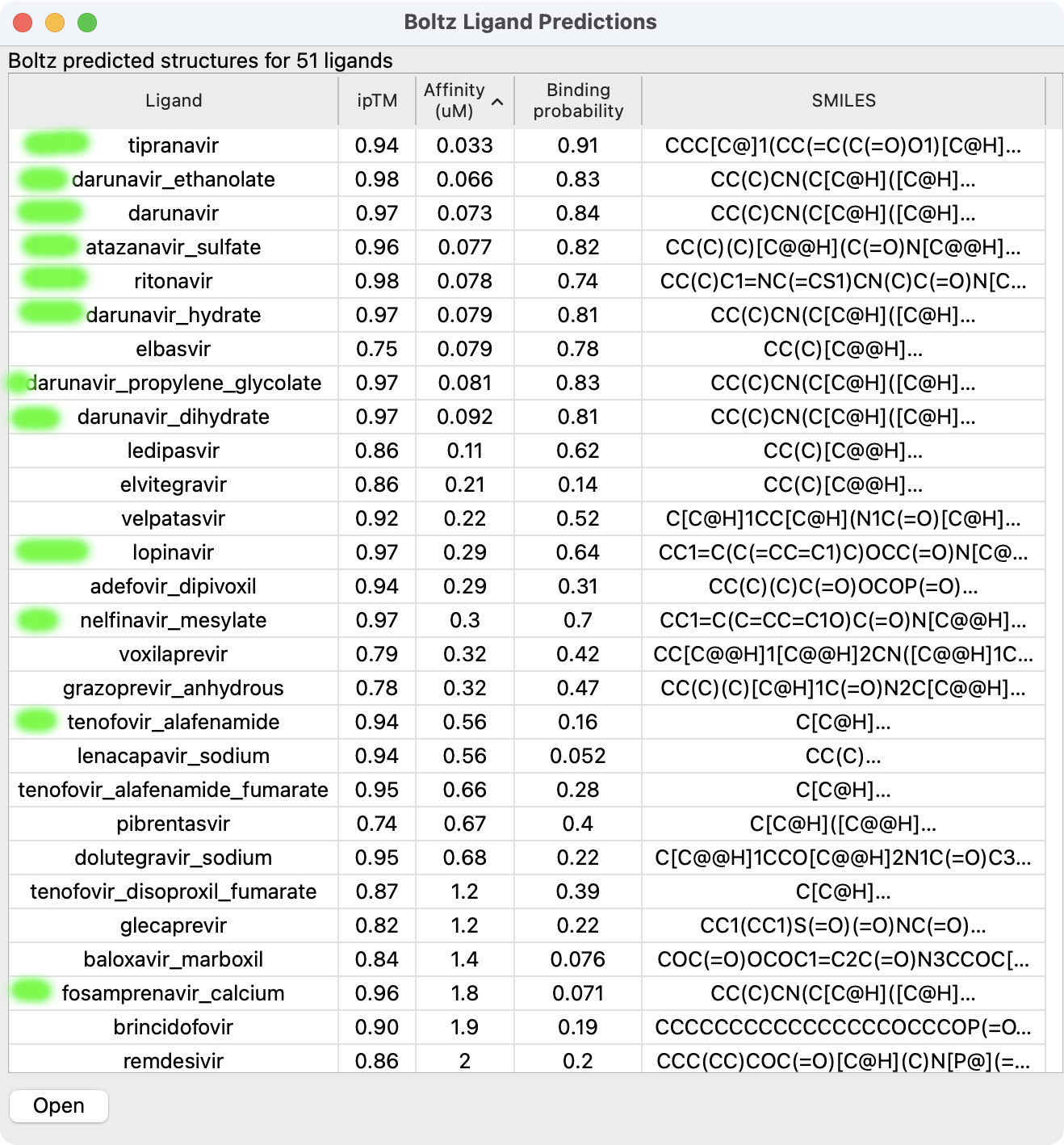
Boltz ligand prediction demo
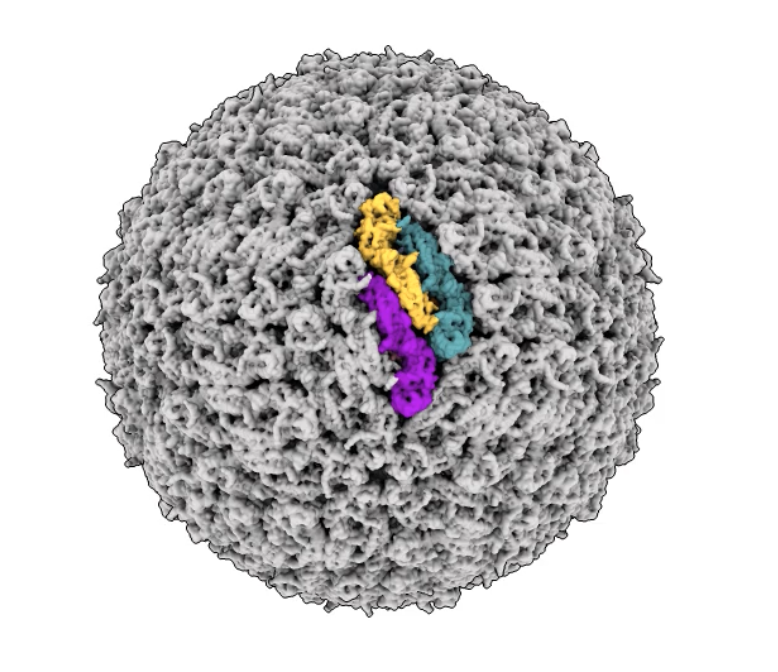
- HIV protease dimer (198 amino acids) docking 51 anti-viral compounds.
- MacBook Pro M1 Max took 2 hours 11 minutes (GPU averaging 30 watts), Linux Nvidia 4090 took 14 minutes (10x faster, GPU averaging 330 watts).
- 12 known hiv protease inhibitors had affinity rankings 1,2,3,4,5,6,8,9,13,18,26 and ipTM rankings 1,2,3,4,5,6,7,8,10,11,15,16.
- HIV protease inhibitors with poorer affinity have high ipTM scores, so combinings scores is more accurate.
When are ligand predictions accurate?
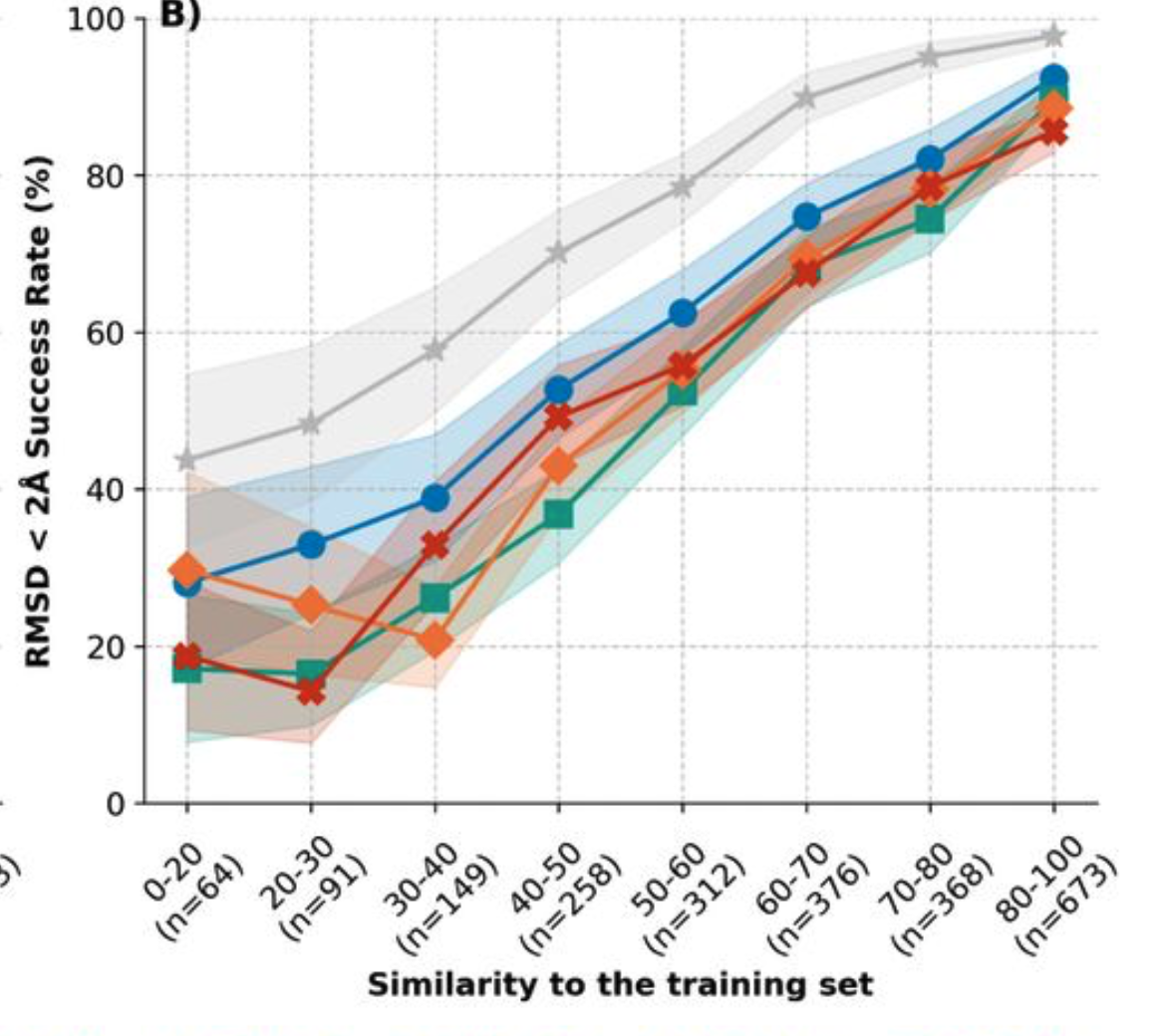
- bioRxiv preprint Have protein-ligand co-folding methods moved beyond memorisation?
- Shows AlphaFold 3, Boltz 2, Chai-1 and Protenix all give ligand accuracy strictly proportional to how often similar ligands and pockets appeared in their training data.
- Experimental ligand binding training data is not extensive enough to generalize to novel ligands an pockets.
- Current methods do seem to summarize well ligand binding seen in training data which is still valuable.
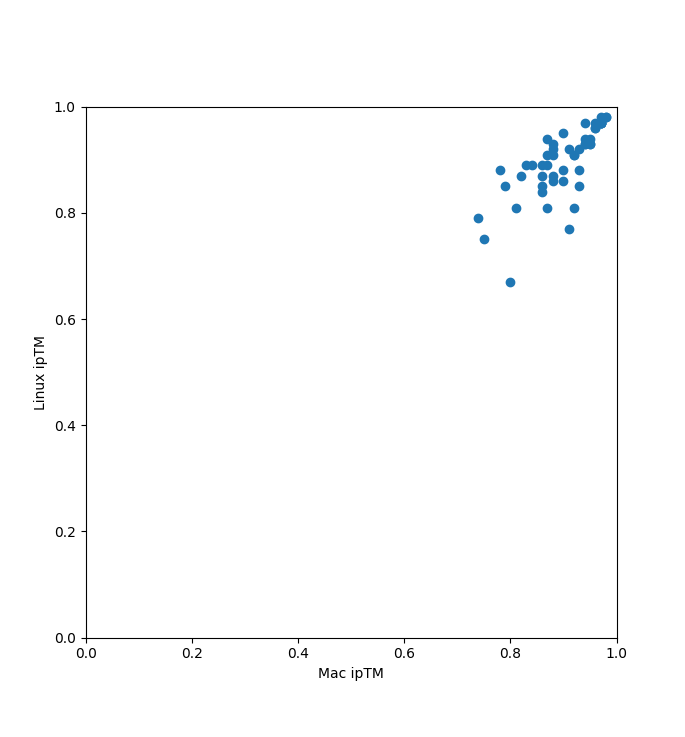
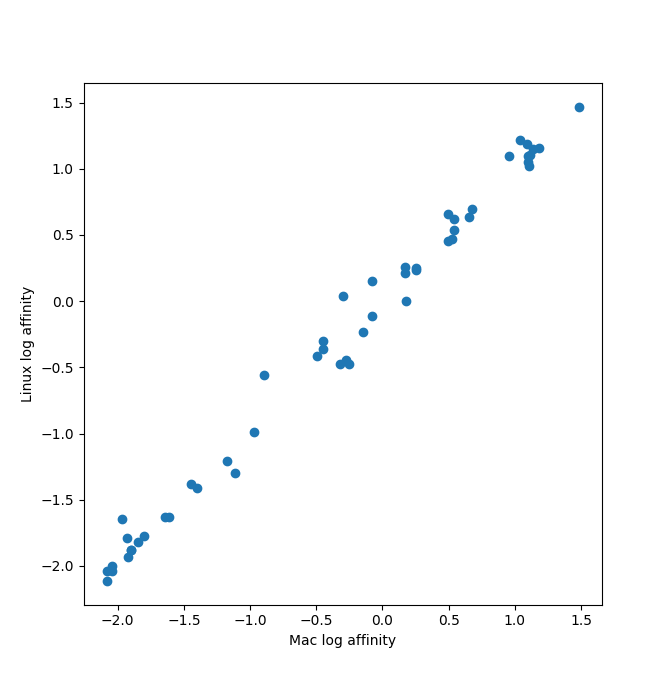
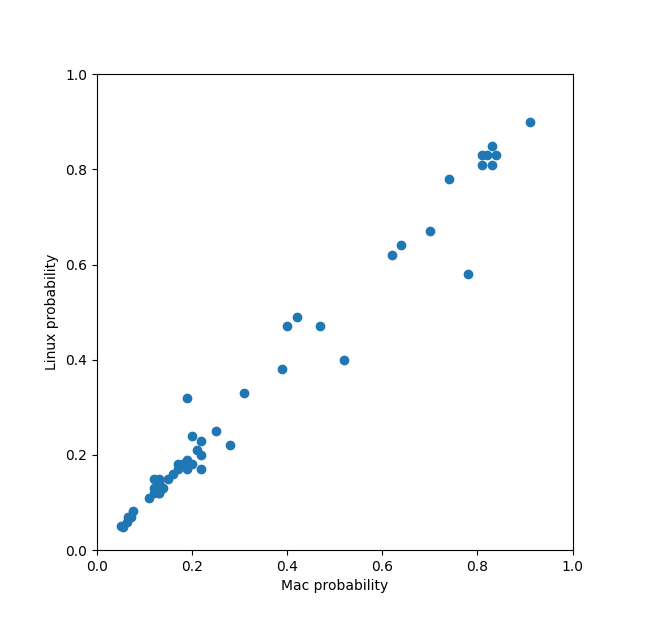
Prediction consistency with different random seeds
The Mac and Linux runs of HIV protease with 51 ligands used different random seeds (based on time) since no
seed was set. These plots compare the ipTM, log binding affinity, and binding probability for the two runs
for the 51 ligands. All dots should be exactly on the diagonal if both runs gave identical results.
Future ChimeraX Boltz improvements
- User interfaces to specify post-translational modifications (e.g. phosphorylated residues), binding pockets, distance constraints, templates, ligands given as atomic structure instead of SMILES or CCD code.
- Improve memory efficiency of Boltz to allow running larger predictions (> 1000 amino acids).
- Allow local MSA calculation using mmseqs.
- ChimeraX Boltz improvements ticket
Animation Examples
Look at example animations we would like to be able to make in an Animation GUI.
These are better than average movies from 2024 Science and Nature papers found by Elaine
to help Tom Goddard find an example to use for a cryoEM animation tutorial in October 2025 at Stanford.
Elaine example. Spinning, morphing, labels, good use of color.
- Elaine movie. Spinning, fading, morphing, labels, 71 seconds.
- Minimal motions.
- Labels match structure colors.
- Worked on for 1 month.
| 
|
- Atomic-resolution structure of a chimeric Powassan tick-borne flavivirus. Paper.
Movie 1, 2.
- Labels match structure colors.
- Atom spheres appear low resolution.
- Models fill little of frame.
| 
|
- Good ambient lighting.
- Spin is too fast, better to rock.
- Swooping motions.
| 
|
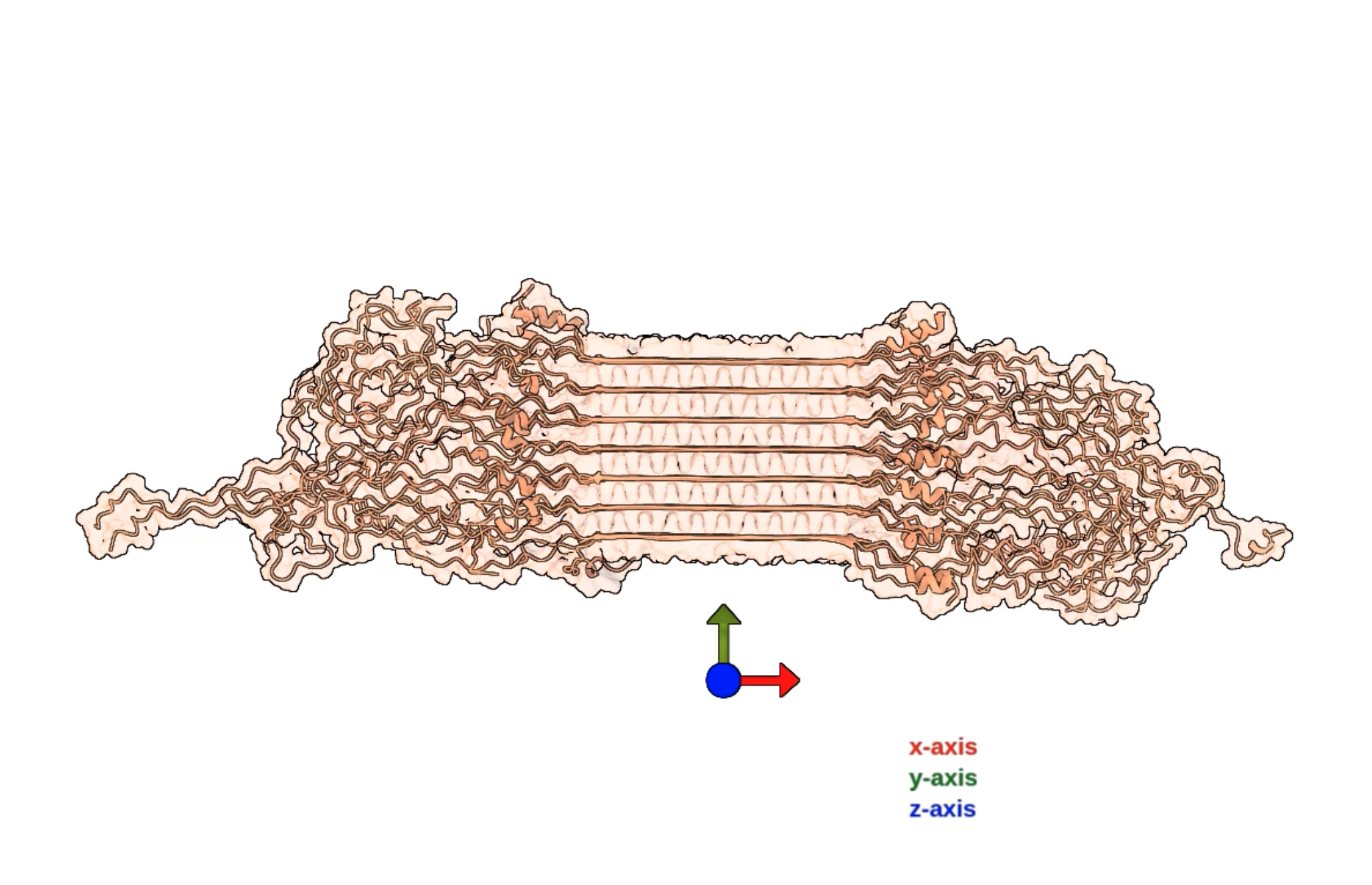
- Integrative determination of atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington disease. Paper.
Movie 1
- Simple montions, map and atomic model.
- 3D labels obscured by atoms.
- Flickering lighting.
| 
|
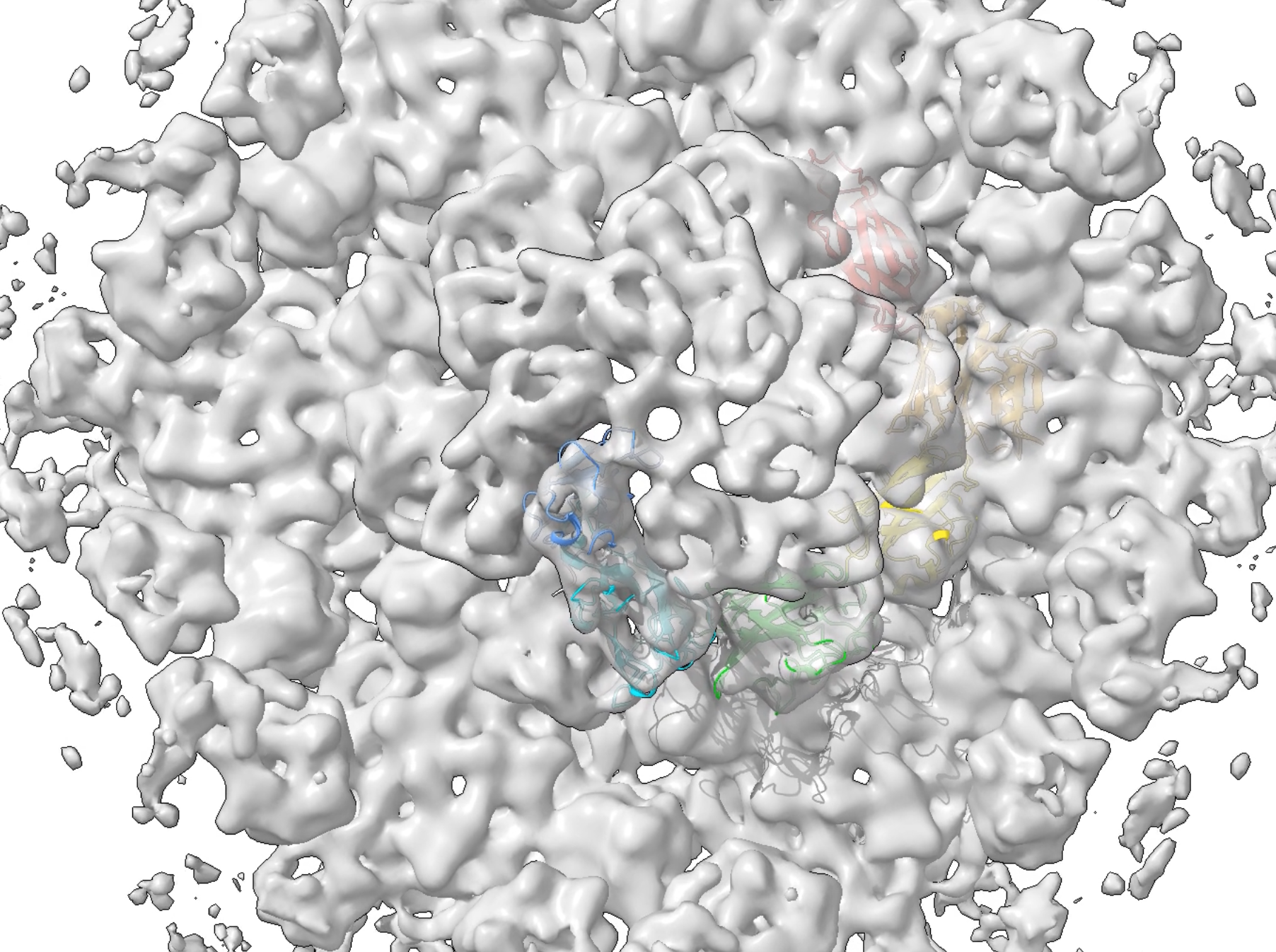
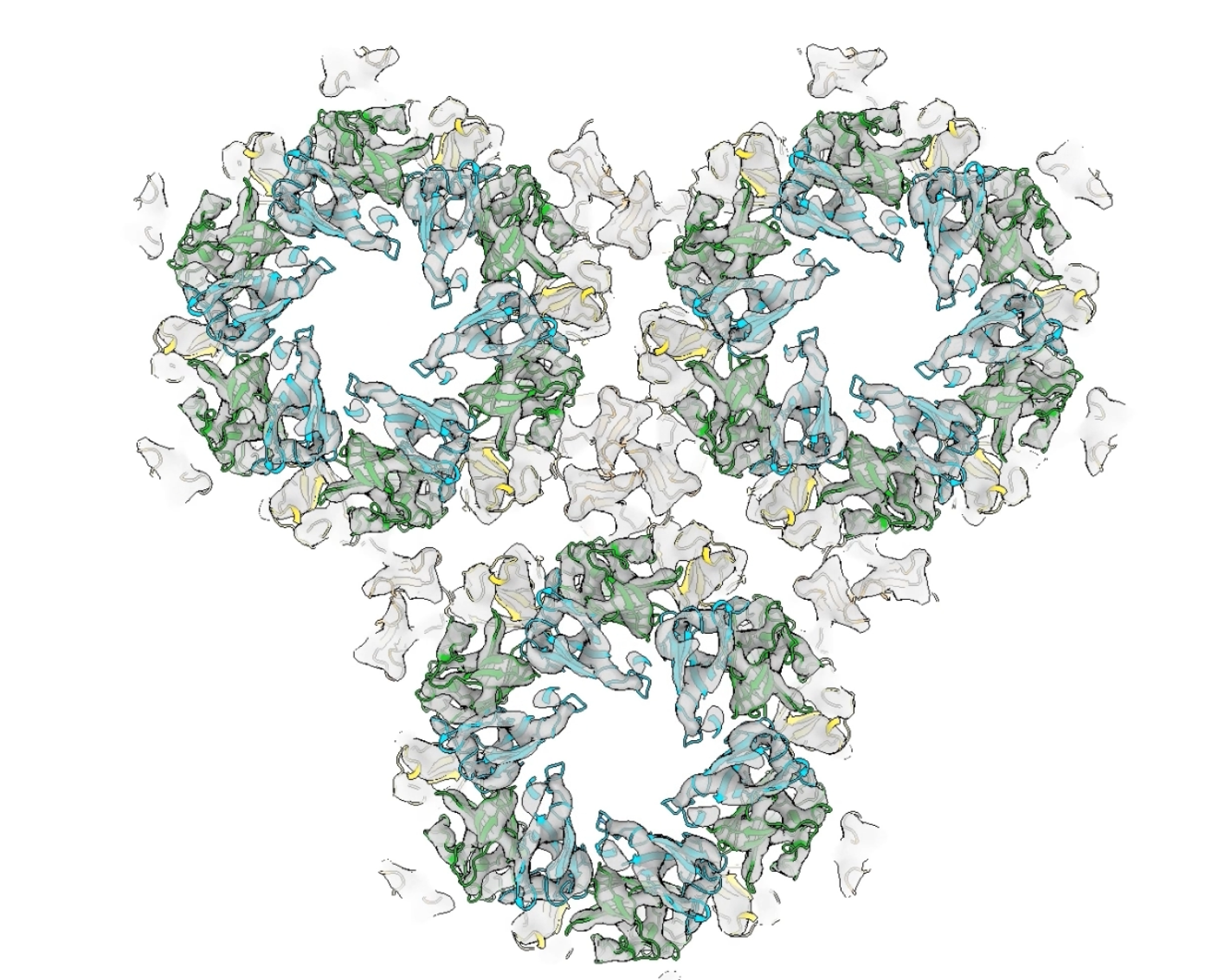
- Composition and in situ structure of the Methanospirillum hungatei cell envelope and surface layer. Paper.
Movie 1,
2,
3.
- Loss of ambient lighting with transparency.
| 
|
| 
|
| 
|
- Molecular sociology of virus-induced cellular condensates supporting reovirus assembly and replication.
Paper.
Movie 4, 5.
- Morph with molecular surfaces.
- Good ambient lighting.
| 
|
- Large scale morph.
- Good colors to distinguish subunits.
| 
|
- Atomic structures of a bacteriocin targeting Gram-positive bacteria. Paper.
Movie 1, 2, 3, 4, 5.
- Cutting open colored EM map.
| 
|
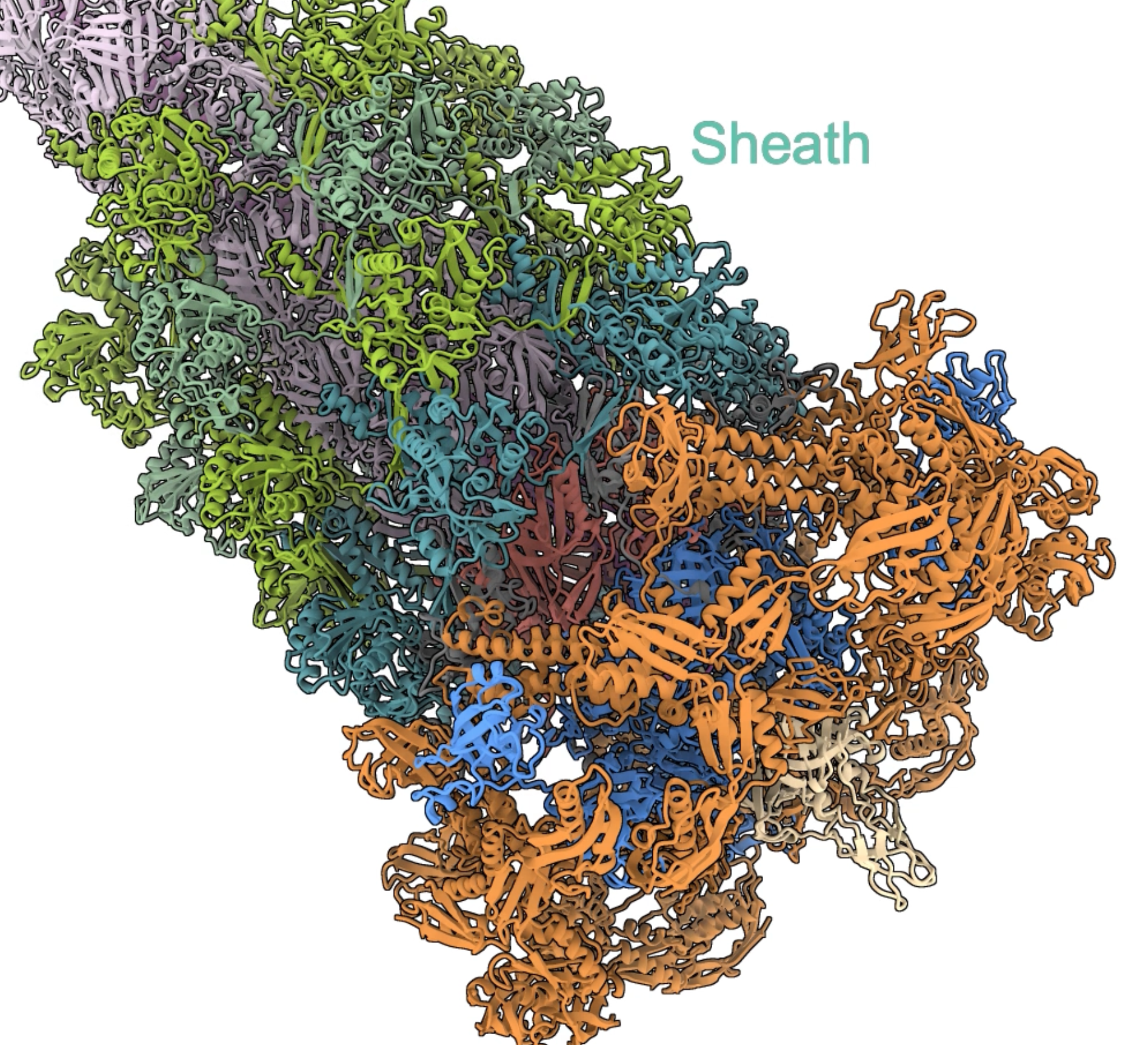
- Fading in many atomic models.
| 
|
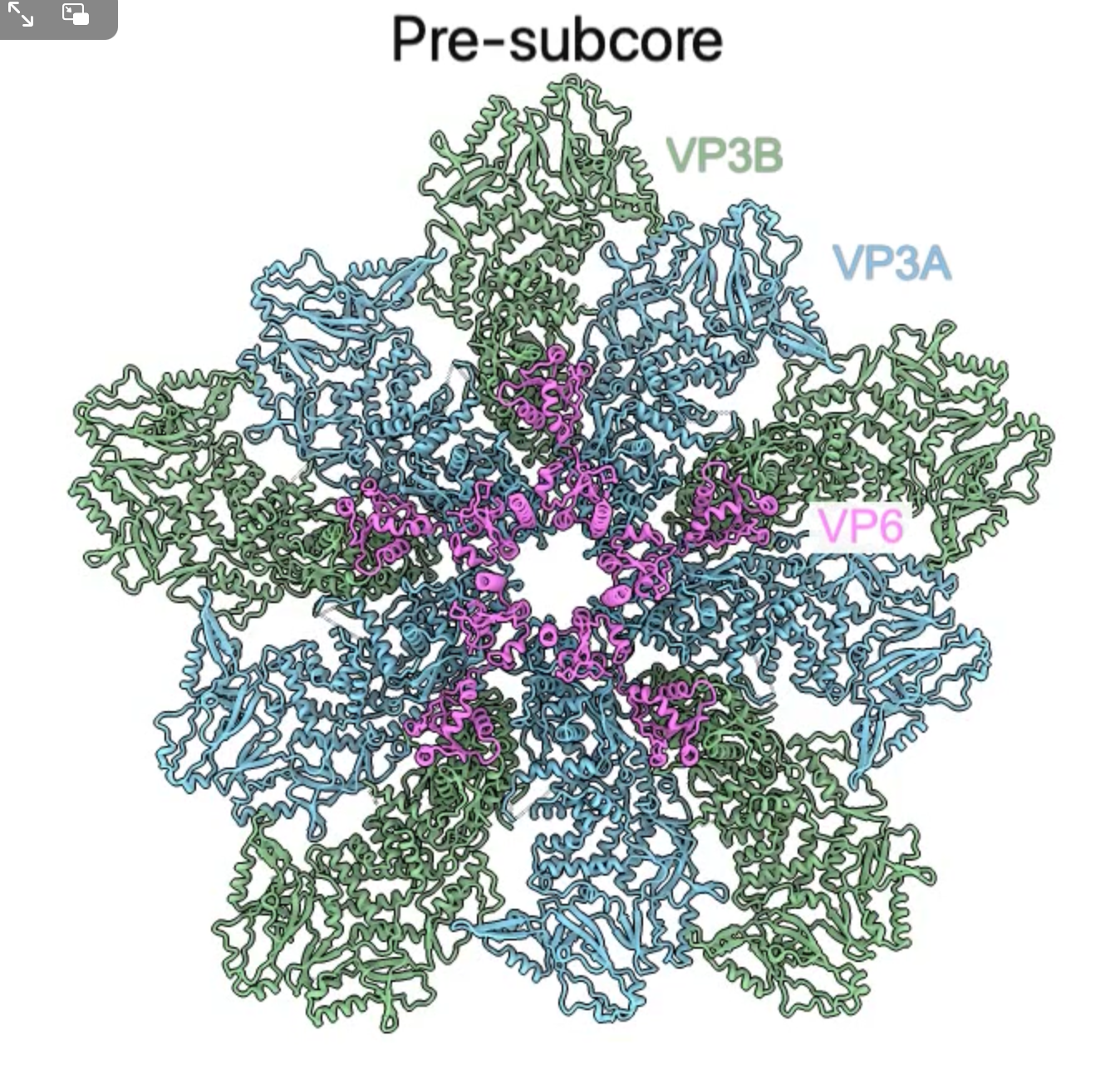
- Comparing and rocking 2 colored maps.
| 
|
| 
|
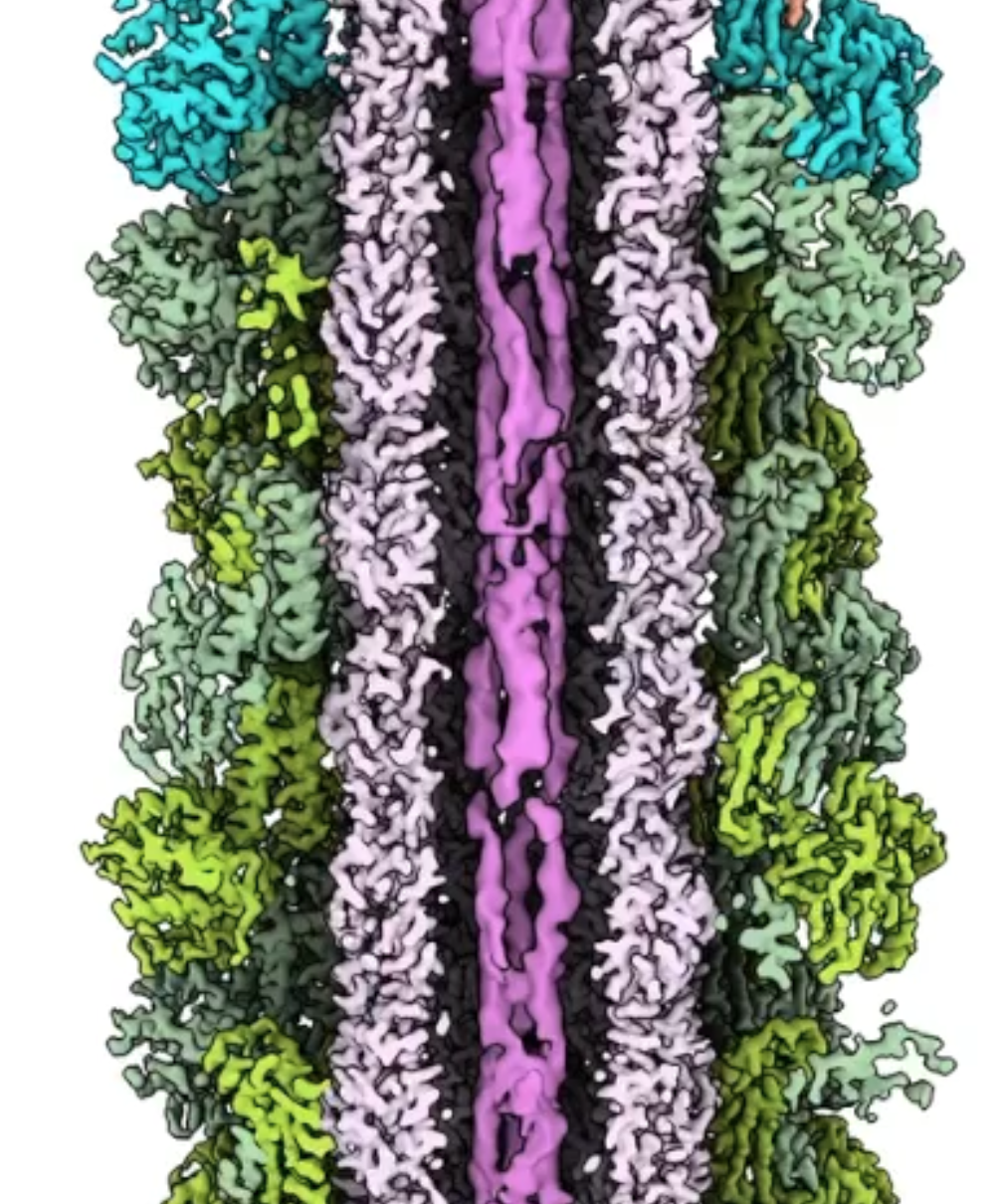
- Panning along sliced open colored filament map.
| 
|
|
- RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells. Paper.
Movie 1, 2, 3, 4.
- Flipping through tomogram planes, then rocking segmented colored structures.
| 
| 
|
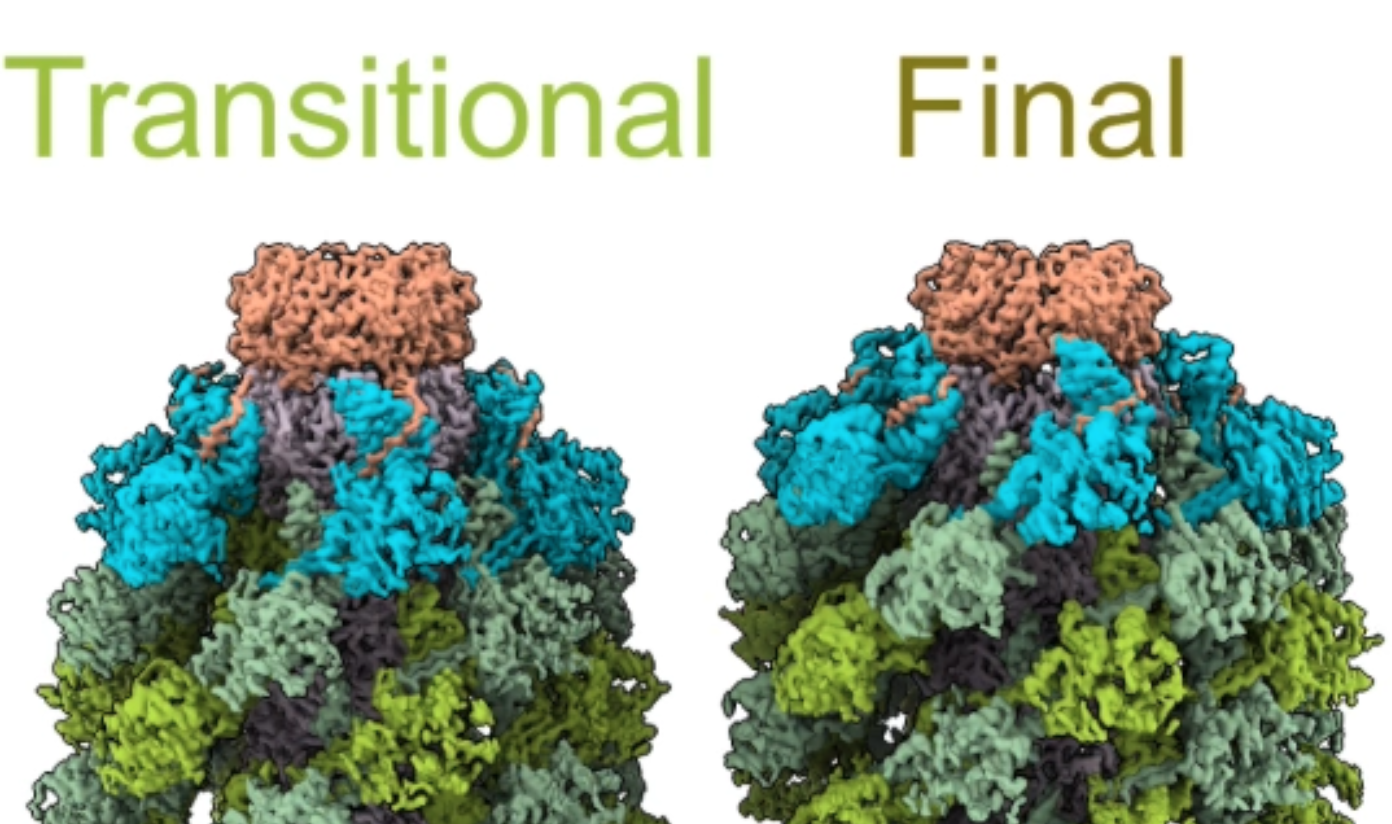
- Morphing large colored atomic model.
| 
| 
|
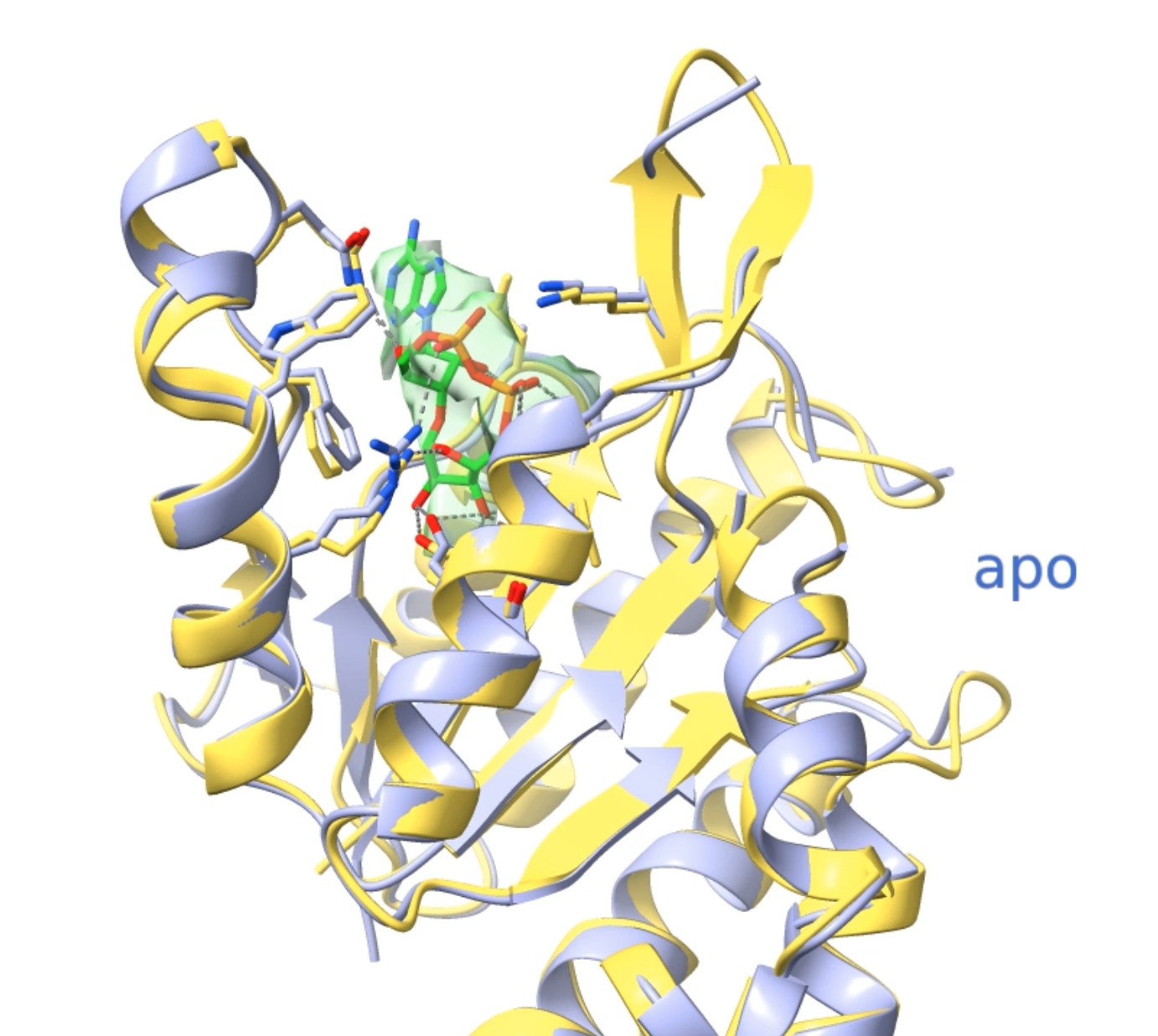
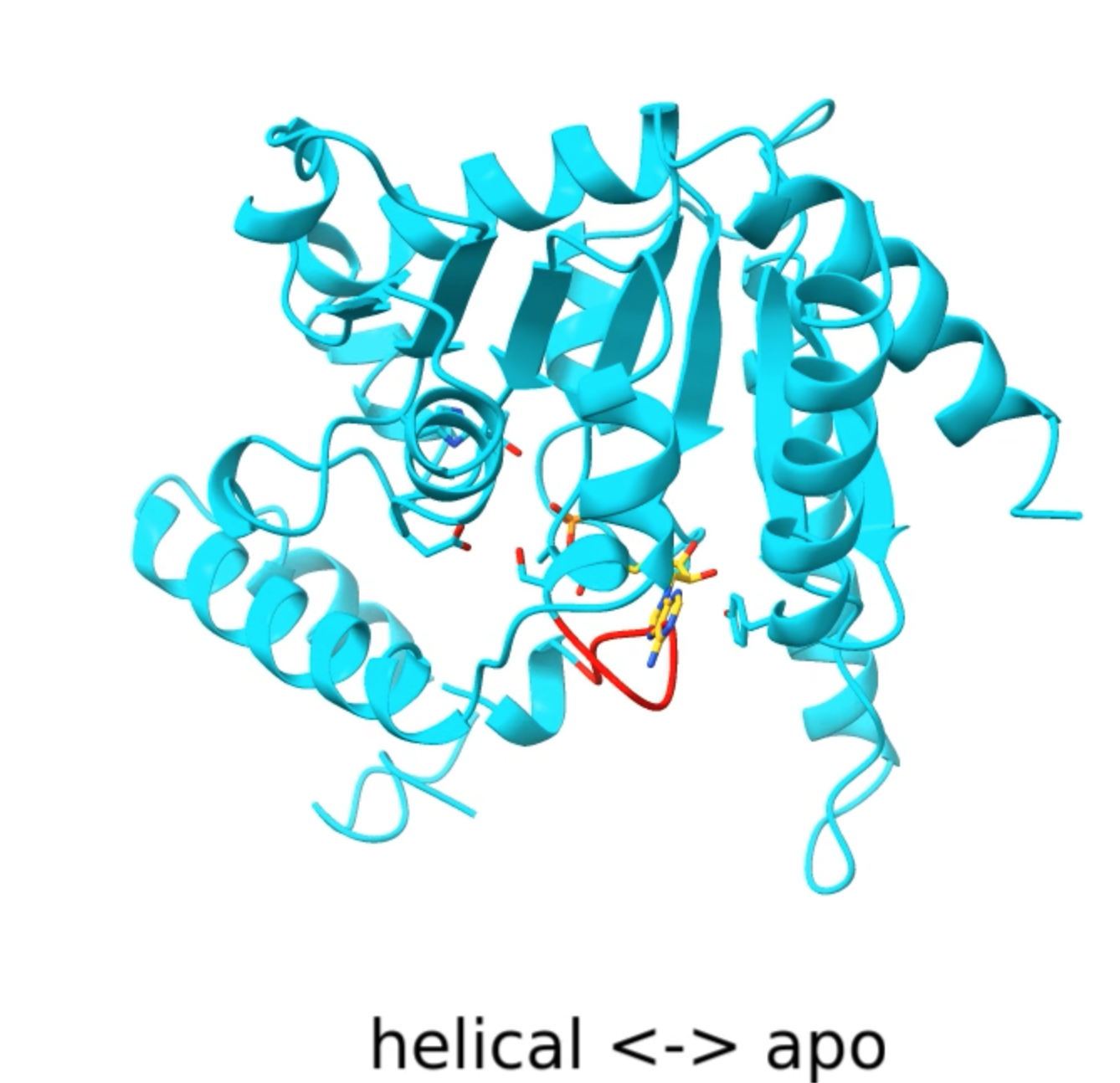
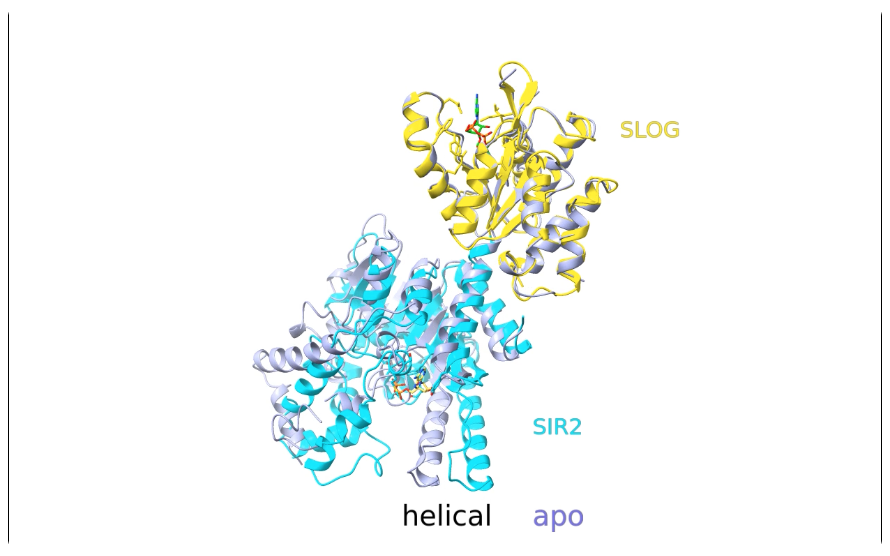
- Activation of Thoeris antiviral system via SIR2 effector filament assembly. Paper.
Movie 1, 2, 3, 4, 5.
- Coloring, labeling and rocking map proteins.
| 
|
- Superimposed atomic models with transparent maps and rocking.
- Odd jump in zoom.
| 
|
- Superimposed atomic models with labels and morphing.
| 
| 
| 
|
Animation Tips
- Don't rotate and translate or zoom at same time.
It makes it hard for the viewer to maintain focus on what they were looking at. Also causes swooping motions that are nauseating.
Example swooping (17 sec mark).
- Rock is usually better than roll.
Rolling 360 degrees usually provides a lot of uninformative views that fly by too fast.
Example roll (5 sec mark),
rock (25 sec mark).
- Put labels close to what they label.
Labels off in the corner of the frame are easily missed when focused on the structures.
Example close (25 sec mark),
close,
far (10 sec mark),
too close (78 sec mark).
- Use label colors that match color of object being labeled.
Example matching.
- Label start and end state of morph.
For example, Apo vs Bound.
Example labeled (12 sec mark).
- 25 frames per second flickers, use 60 frames per second.
Example 25 fps, 60 fps.
- Maintain ambient shadows on transparent surfaces.
Gives better 3d appearance. Switch from ambient shadows to no shadows when making surface transparent is jarring. Requires tricky change to surface material to cast shadows and make structures inside not receive shadows.
Example no shadows (6 sec mark),
no shadows (3 sec mark).
- Make models fill most of frame.
Big frame with small models makes detail hard to see.
Example too small.