
Tom Goddard
Updated December 15, 2025
ChimeraX can run the Boltz structure prediction method to compute atomic structures of proteins and nucleic acids, including modified residues, ligands, ions and solvent on your laptop or desktop computer. ChimeraX daily builds dated July 22, 2025 and newer use Boltz 2.1.1 and include ligand affinity prediction. ChimeraX 1.10 released June 26, 2025 uses Boltz 1.0.0. See the demonstration video or affinity prediction video or batch ligands video. The method is inspired by AlphaFold 3 and described in the following preprint and is fully open source with a very permissive MIT license.
Boltz-1: Democratizing Biomolecular Interaction Modeling
Jeremy Wohlwend, Gabriele Corso, Saro Passaro, Mateo Reveiz, Ken Leidal,
Wojtek Swiderski, Tally Portnoi, Itamar Chinn, Jacob Silterra, Tommi Jaakkola, Regina Barzilay
bioRxiv 2024.11.19.624167; doi: https://doi.org/10.1101/2024.11.19.624167
A ChimeraX graphical user interface (menu Tools / Structure Prediction / Boltz) and ChimeraX command (boltz) are provided to make predictions. The latest version of this documentation is available online.
Boltz structure predictions use a lot of memory and compute resources on your computer that limits the size of the structure that can be predicted. See the run times section below for example run times and size limits.
Mac. It works well on Mac M-series (M1,M2,M3,M4) laptop and desktop computers predicting small 100 residue structures in 1 minute and up to 1200 residues in about 15 minutes with 32 GB of memory. With 16 GB of memory it can only predict about 350 amino acids taking about 5 minutes, with larger predictions running out of memory.
Nvidia GPUs on Windows and Linux. Nvidia GPUs on Windows and Linux computers also provide good performance. On Linux with an Nvidia GPU with 24 GB of graphics memory (e.g. Nvidia RTX 3090 or 4090) it can predict about 1300 residues in about 4 minutes, larger sizes run out of memory. Testing on Windows with less GPU memory, 12 GB (e.g. Nvidia RTX 4070) predicts up to 1000 residues. On Windows with 8 GB of GPU memory (e.g Nvidia RTX 3070) predictions of about 700 residues. Large predictions on Nvidia GPUs on Linux run out of memory and fail. On Windows the prediction will fallback to using CPU memory allowing larger structure predictions but taking immensely longer run times (10-30 times longer).
Intel CPU. Predictions only utilizing an Intel CPU are very slow, for example 1.5 hours for 900 residues. Expected size limits are about 350 residues with 16 GB, 1000 residues with 32 GB, and about 1600 residues with 64 GB. Run time is expected to increase as the square of the number of residues.
When you first start the Boltz tool within ChimeraX (menu Tools / Structure Prediction / Boltz) it will show a button Install Boltz. Boltz is a large software package, taking about 1 Gbyte of disk space and uses the Torch machine learning package. It also requires the neural network weights (3.3 Gbytes) and PDB Chemical Component Dictionary (CCD, 0.3 Gbytes) to make predictions. Downloading and installing all of these files will take 10 minutes or more depending on network speed and install the software in your home directory in ~/boltz2 and the network weights and CCD in ~/.boltz.
The ChimeraX boltz install command can also be used to do this one time installation.

|
Menu of molecular components
|
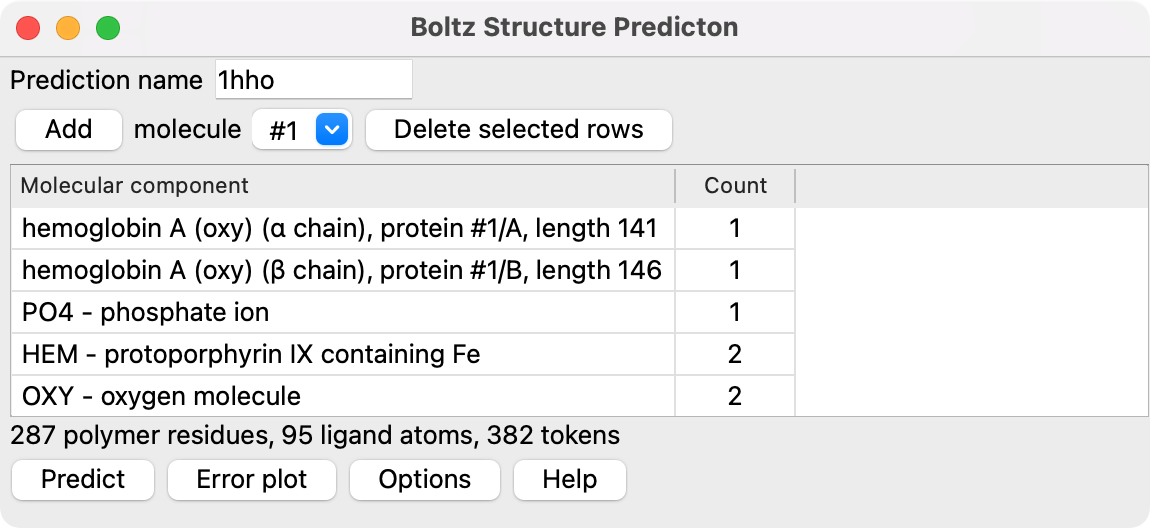
To predict a structure made up of proteins, nucleic acids and small molecules you first specify all the molecular components. Choose entries from the Add menu and press the Add button to add them to your assembly specification in the table below. You can specify component molecules in several ways.

Hemoglobin dimer prediction (blue) aligned to experimental structure PDB 1hho |
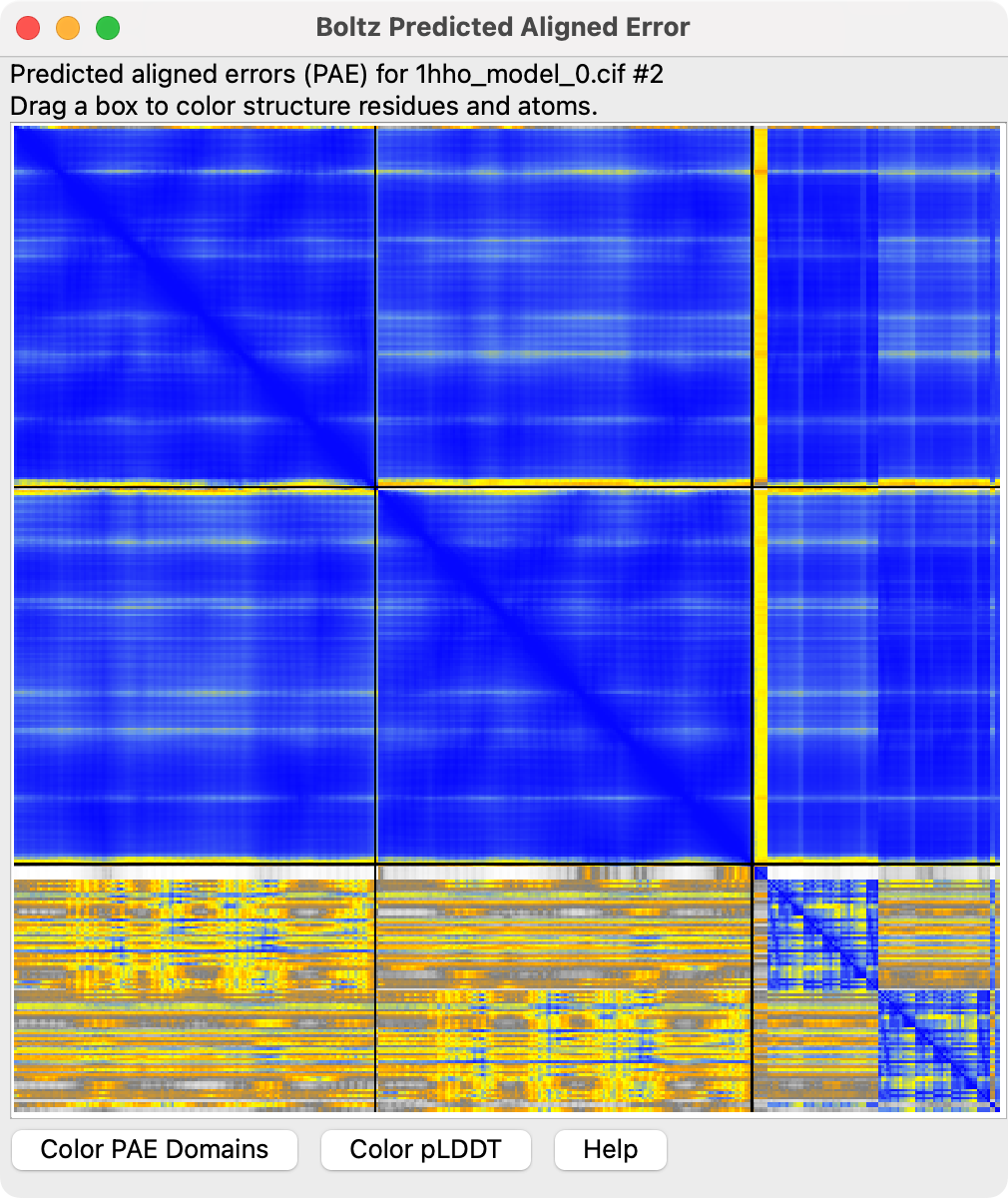
Predicted aligned error |
Components can be added multiple times to have more instances of that molecule in the assembly. Press the Predict button after the assembly is completed by adding each component to start the prediction. A Stop button will be shown while the prediction runs to terminate the prediction, discarding the partial computation so you can start another prediction.
The results are put in a new folder on your desktop named by default boltz_assembly_name where the prediction name can be specified at the top of the ChimeraX Boltz panel. Using the Options described below you can change where the result files are placed.
Predictions for small assemblies, for example 500 residues and ligand atoms, take one to several minutes depending on the computer (e.g. Nvidia GPU vs CPU only). The run in the background (a separate process) so ChimeraX can continue to be used while the calculation runs. The predicted structure will be opened in ChimeraX when the calculation completes. If the assembly specification involved proteins or nucleic acids specified using chains of open models, the predicted structure will be aligned (using matchmaker) to the open model for the first such component.
The prediction will be colored using the standard AlphaFold pLDDT type of coloring where blue indicates high confidence, yellow and red moderate to low confidence.
Finer grained estimates of prediction confidence can be displayed by pressing the Error Plot button to show the predicted aligned error.
Here are run times for several desktop and laptop computers for predicting various size molecular assemblies from the Protein Databank using Boltz version 2.1.1 (July 9, 2025 source). Another table compares Boltz 2.1.1 to 1.0.0 to 0.4.1 and timings of 16-bit vs 32-bit floating point, diffusion steering, and affinity prediction.
Boltz prediction times in minutes. Tokens is number of standard polymer residues plus ligand atoms.
| PDB code | Tokens | Mac M1 16 GB | Mac M1 Max 32 GB | Mac M2 Ultra 64 GB | Linux i9 CPU 64 GB | Linux Nvidia 4090 | Windows i7 CPU 64 GB | Windows Nvidia 3070 | Number of residues and atoms and prediction error |
|---|---|---|---|---|---|---|---|---|---|
| 8rf4 | 129 | 1.2 | 0.8 | 0.8 | 1.1 | 0.3 | 2.0 | 0.8 | 118 amino acids, 11 ligand atoms, 0.8A RMSD 108 residues |
| 1hho | 382 | fail | 1.5 | 1.4 | 8.0 | 0.5 | 11 | 2.0 | 287 amino acids, 95 ligand atoms, 0.4A RMSD 146 residues |
| 9moj | 660 | 18 | 3.8 | 2.3 | 24 | 0.6 | 29 | 2.5 | 660 amino acids, heterotetramer, 0.8A RMSD 132 residues |
| 9h1k | 671 | 23 | 4.9 | 2.7 | 29 | 0.7 | 38 | 9.5 | 560 amino acids, 59 rna bases, 52 ligand atoms, 0.7A RMSD protein, RNA wrong |
| 9b3h | 911 | fail | 10 | 5.1 | 60 | 1.1 | 80 | 28 | 911 amino acids, heterodimer, 1A RMSD 509 residues |
| 9fz5 | 1025 | 13 | 7.0 | 82 | 1.3 | 40 | 1025 amino acids, heterotrimer, 3.7A RMSD 740 residues | ||
| 9mcw | 1154 | 18 | 7.2 | 96 | 1.4 | 54 | 1154 rna bases, homodimer, wrong dimer and monomer conformations | ||
| 8sa0 | 1371 | 28 | 38 | 164 | fail | 1274 amino acids, 97 ligand atoms, 3.4A RMSD 1151 residues | |||
| 9gh4 | 1467 | 32 | 41 | 188 | 2.1 | Protein homotrimer, monomer 489 residues, 1.1A RMSD for 750 residues |
Mac GPU acceleration. The reported Mac performance is for Mac M1/M2/M3/M4 series GPUs. Boltz uses machine learning package torch which has GPU acceleration called Metal Performance Shaders (MPS) on these Mac M series GPUs which have speed up to 2-3x slower than an Nvidia 4090 but with the advantage that the Mac can handle larger molecular systems using the unified computer memory (e.g. 32 or 64 GB). With 16 GB prediction size is limited to 350 residues. Older Mac Intel machines do not have GPU acceleration in Torch and run at speeds similar to Windows Intel CPU-only predictions.
Windows Nvidia GPU performance. The above table shows a significant slow-down in predictions beyond about 600 residues on Windows with Nvidia 3070 (8 GB) and 4070 (12 GB) graphics. This is probably because the GPU memory is insufficient for larger structures and the machine learning toolkit falls back to a mix of CPU and GPU calculation. Notice that the 4070 GPU took more time than the 3070 GPU for large structures probably because the CPU on the 4070 machine (i5-6700K) is significantly slower than the CPU on the 3070 machine (i7-12900K).
Linux Nvidia GPU out of memory. On Linux Nvidia 4090 with 24 GB of GPU memory the maximum prediction appears to be about 1300 residues plus ligand atoms before an "out of memory" error occurs. This contrasts with Windows where Torch appears to fallback to using CPU and not run out of GPU memory.
Optimizing for larger structures with Nvidia GPU. Structure predictions 40% larger are possible by using 16-bit floating point (bfloat16) on Nvidia GPUs instead of float32 for boltz weights and activations. Prediction speed is about the same. I tested this on Nvidia 4090 on Linux and it was able to predict 9fz5 (1025 tokens) in 3 minutes and 8sa0 (1371 tokens) in 5 minutes. I have not seen a reduction in accuracy. Details in ChimeraX ticket #17555. Boltz 2 makes use of 16-bit floating point (bf16-mixed) by default when using Nvidia graphics. Mac and Intel hardware give worse speed and little memory savings with 16-bit because the Torch 2.9 package does not support native 16-bit on those architectures and instead uses 32-bit float.
Tom Litfin made Boltz optimizations to reduce GPU memory use so that larger predictions can be made. The main optimization moves some Torch tensors from GPU memory to CPU memory as described in LMI4Boltz: Optimising VRAM utilisation to predict large macromolecular complexes with consumer grade hardware (November 2025). Another optimization changes Torch tensors in place instead of making copies. And a third optimization allows different "chunking" in the neural net attention layers that reduces memory use. These optimizations increase the number of residues that can be predicted on an Nvidia 4090 GPU (24 GB) with 64 GB of CPU memory from 1300 to 2200 as shown in the following table. With chunking optimization up to 2500 residues can be predicted. It also can reduce the prediction times on Windows with Nvidia GPUs with less memory (e.g. 8 GB or 12 GB).
Low memory Boltz prediction times in minutes. Tokens is number of standard polymer residues plus ligand atoms.
| PDB code | Tokens | Linux Nvidia 4090 | Linux Nvidia 4090 Low Mem | Windows Nvidia 3070 | Windows Nvidia 3070 Low Mem | Number of residues and atoms and prediction error |
|---|---|---|---|---|---|---|
| 8rf4 | 129 | 0.3 | 0.8 | 118 amino acids, 11 ligand atoms, 0.8A RMSD 108 residues | ||
| 1hho | 382 | 0.5 | 2.0 | 1.5 | 287 amino acids, 95 ligand atoms, 0.4A RMSD 146 residues | |
| 9moj | 660 | 0.6 | 2.5 | 2.5 | 660 amino acids, heterotetramer, 0.8A RMSD 132 residues | |
| 9h1k | 671 | 0.7 | 9.5 | 5.5 | 560 amino acids, 59 rna bases, 52 ligand atoms, 0.7A RMSD protein, RNA wrong | |
| 9b3h | 911 | 1.1 | 28 | 10 | 911 amino acids, heterodimer, 1A RMSD 509 residues | |
| 9fz5 | 1025 | 1.3 | 40 | 19 | 1025 amino acids, heterotrimer, 3.7A RMSD 740 residues | |
| 9mcw | 1154 | 1.4 | 54 | 1154 rna bases, homodimer, wrong dimer and monomer conformations | ||
| 8sa0 | 1371 | failed | 2.6 | 101 | 1274 amino acids, 97 ligand atoms, 2.2A RMSD all residue CA. | |
| 9gh4 | 1467 | 2.1 | 2.8 | Protein homotrimer, monomer 489 residues, 1.1A RMSD for 750 residues | ||
| 8zvq | 1550 | failed | 3.3 | |||
| 9d45 | 1651 | 3.8 | ||||
| 9enr | 1794 | 3.9 | ||||
| 1dpp | 2028 | 5.4 | ||||
| 1a4e | 2146 | 6.1 | ||||
| 9hma | 2217 | 7.1 | ||||
| 7r76 | 2348 | 9.5 | Used --aggressive_chunking. Out of memory without this option. | |||
| 7lhw | 2527 | 11 | Used --aggressive_chunking. | |||
| 7b52 | 2649 | failed | Used --aggressive_chunking. |
Here are few low memory boltz tests on GPUs with more memory (without aggressive chunking, tested on RunPod.io).
7lhw, 2527 residues, Nvidia 5090 (32 GB), 7 minutes
7lhw, 2527 residues, Nvidia L40S (48 GB), 12 minutes
8p52, 2971 residues, Nvidia L40S (48 GB), 21 minutes
This low memory Boltz should give the same predictions as regular Boltz. It has not yet been incorporated into the main Boltz 2 github repository (Dec 2025), but there is a special ChimeraX version of Boltz that uses the low memory version. To install and use the low memory Boltz use ChimeraX command
boltz install ~/boltz22_lowmem branch chimerax_boltz22_lowmem
ChimeraX currently currently does not use the "aggressive chunking" option.
Several predictions can be run each with a different ligand to see how it binds to the rest of the molecular assembly. Use the Add molecule "each ligand SMILES string" menu entry. Paste in a set of SMILES strings with ligand names. Each line should have a ligand name, comma, followed by a SMILES string. The ligand names will be used as the predicted structure file names. Alternatively you can just enter one SMILES string per line without a name and the ligands will be named "ligand1", "ligand2", .... Press the Add button after pasting in the set of ligands, then the Predict button to run the series of predictions.
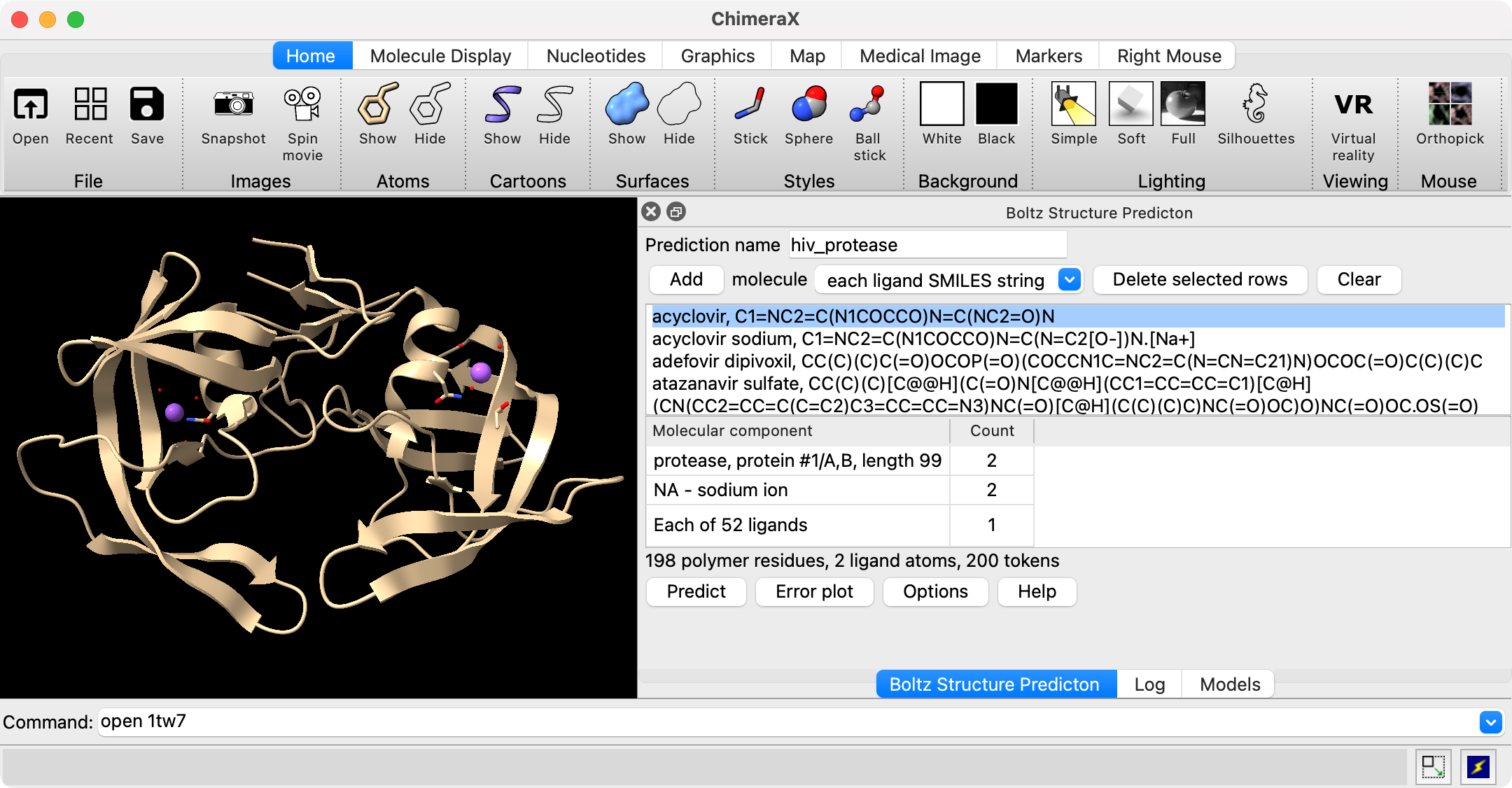
Example input for 52 anti-viral ligands run against HIV protease. Predictions completed in 14 minutes on an Nvidia 4090 GPU. | 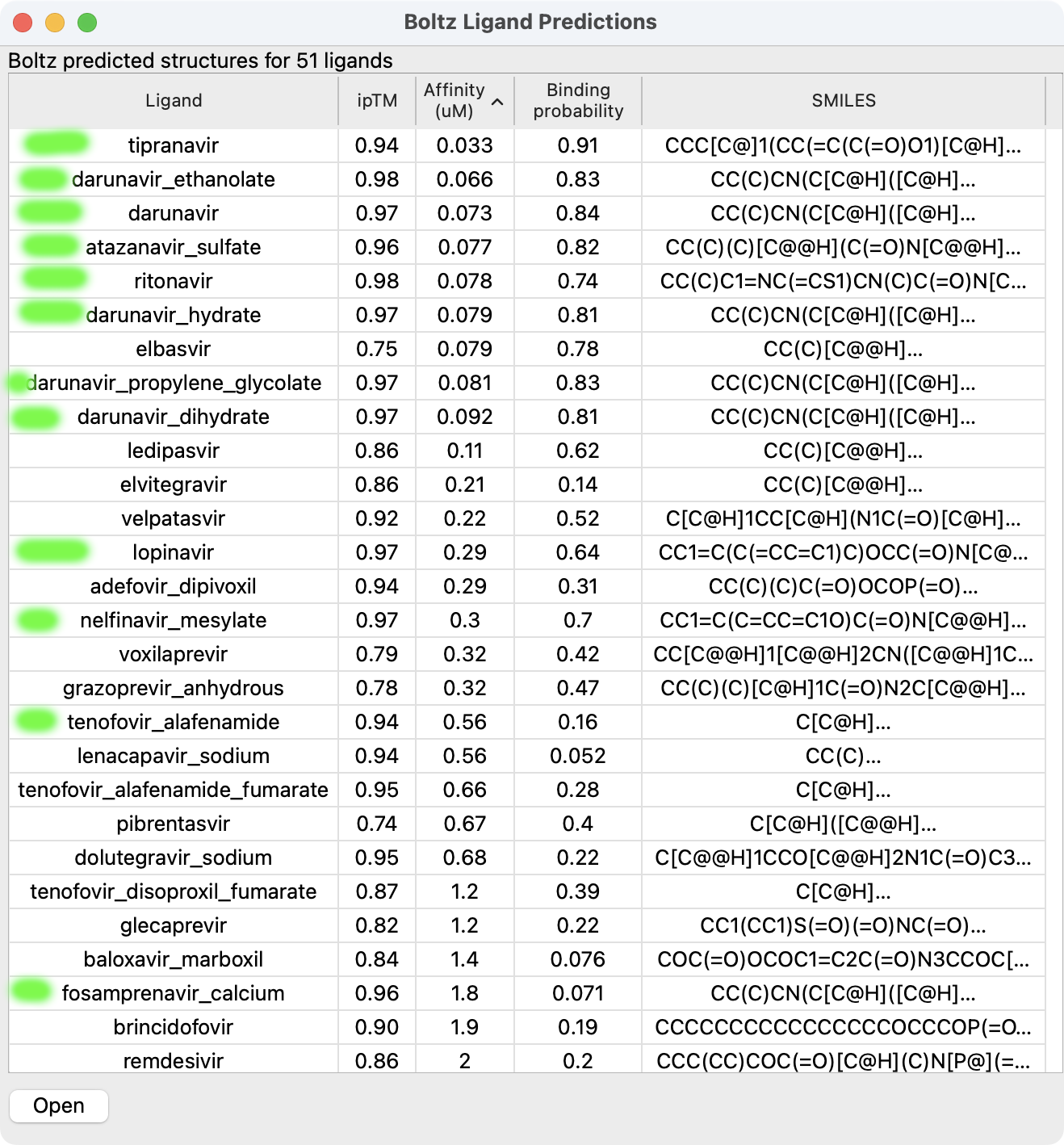
Binding confidence and affinity scores for ligands. Green highlights shows known HIV protease inhibitors. |
Progress messages will appear in the Boltz panel as the structures are predicted for each ligand. After all structures are predicted the binding affinity is predicted. About half of the time is taken to predict affinity and if you don't want affinity scores you can set "Predict ligand binding affinity: none" under Options to speed up the calculation. When the predictions complete a table of results giving ligand name, ipTM, affinity, binding probability, and SMILES string will appear. The table can be sorted by clicking on the column headers. Selecting rows of the table and pressing the Open button will open the selected structure predictions. The table is written as comma-separated values to the directory where Boltz was run, for example hiv_protease/hiv_protease.bzlig in the case shown. This file will appear in the file history thumbnails when you start ChimeraX in future sessions and clicking on the thumbnail will reshow the table.

Pressing the Options button shows additional settings for boltz predictions.
Additional advanced options are available by using the ChimeraX boltz command.
|
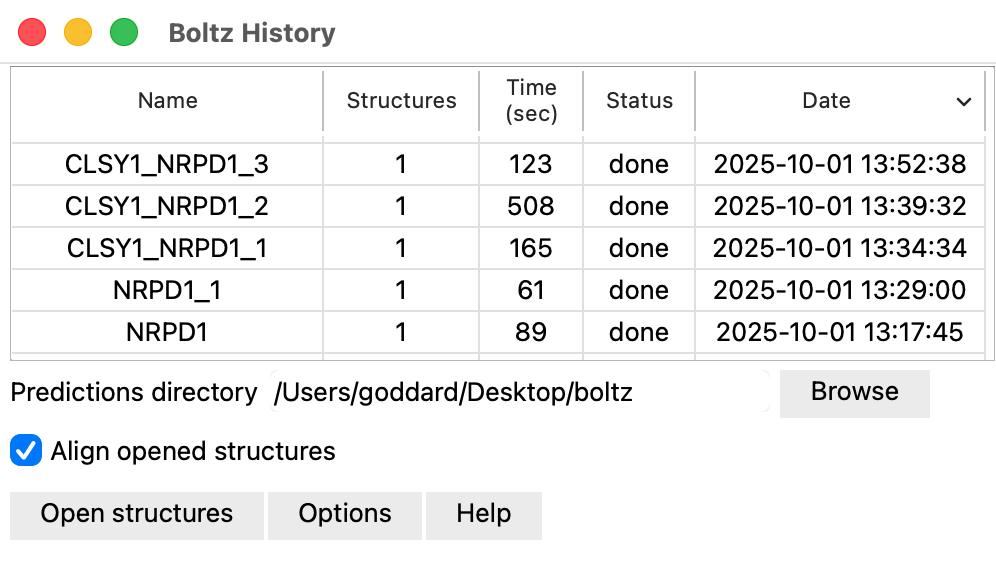
ChimeraX menu entry Tools / Structure Prediction / Boltz History lists a table of past Boltz predictions. Selecting rows of the table and pressing the Open structures button will open those structures. The table is filled by scanning the Boltz prediction directories located in the directory where ChimeraX puts new Boltz predictions (by default ~/Deskop). If a prediction computed multiple structures they will all be aligned to the first structure of that prediction (using the ChimeraX matchmaker command). The directory containing the prediction directories and the choice to align can be specified in the panel shown by pressing the Options button.
Directories are identified as containing Boltz predictions if they contain a file named "command". When ChimeraX runs a prediction it saves the boltz command including all its arguments in this file.
Server predictionsIf there are server predictions that did not complete before quitting ChimeraX, then when ChimeraX is run again the Boltz History panel will show a button Fetch from server next to the Open structures button. This button fetch any server predictions that have completed and opens them. Predictions that have not yet completed are noted in the Log after pressing this button.
The ChimeraX Boltz graphical interface runs a prediction by running the ChimeraX boltz command. That command is recorded in the ChimeraX Log panel, and looking at that command can help you understand the command options.
boltz predict [sequences] [protein sequences] [dna sequences] [rna sequences]
[ligands residue-spec] [excludeLigands ccd-codes]
[ligandCcd ccd-codes] [ligandSmiles smiles-string]
[forEachSmilesLigand name,smiles-string,name,smiles-string...]
[affinity ligand-ccd-or-smiles-string | each | none]
[name prediction-name] [resultsDirectory directory]
[device default|cpu|gpu] [useServer true | false] [serverHost hostname] [serverPort port]
[float16 true|false] [steering true|false]
[samples n] [recycles n] [seed n]
[useMsaCache true|false] [msaOnly true|false]
[open true|false] [installLocation directory] [wait true|false]
Options descriptions
boltz install [directory] [downloadModelWeightsAndCcd true | false] branch name
The boltz install command creates a Python virtual environment to install Boltz from PyPi. If no directory is specified then ~/boltz2 in the user's home directory is used. The directory will be created or if it already exists must be empty. It then downloads the Boltz network parameters and Chemical Component Dictionary to ~/.boltz. Finally it makes an index of the atom counts for each CCD code so that the ChimeraX Boltz interface can report the total number of tokens (residues plus ligand atoms) in an assembly in order to judge whether the computer has enough memory to make the requested prediction.
The install uses a fork of the Boltz repository https://github.com/RBVI/boltz. It uses git branch chimerax_boltz22 of this fork unless the branch option is specified in which case it installs the specified branch. The branch option is for testing new versions of Boltz.
The install process executes these commands to make the virtual environment and install Boltz. It uses the ChimeraX Python executable to create the virtual environment. Boltz will no longer work if ChimeraX is moved or deleted and will need to be reinstalled in that case. It will also no longer work if the boltz directory itself is moved since the boltz executable refers to the install location to find python.
The ChimeraX boltz install command creates a Python virtual environment and installs boltz and downloads the boltz weights and CCD database. On Windows it installs a version of torch with CUDA 12.6 support before installing boltz if Nvidia graphics is detected.
python -m venv directory
directory/bin/python -m pip install torch --index-url https://download.pytorch.org/whl/cu126 # On Windows with Nvidia GPU only.
directory/bin/python -m pip install boltz
directory/bin/python chimerax/site-packages/boltz/download_weights_and_ccd.py
boltz ligandtable runDirectory [ includeSmiles true | false ] [ alignTo atomic-model ]
The boltz ligandtable command displays a table for batch ligand predictions. Normally this command is not needed because ChimeraX batch ligand predictions are tabulated in the directory where the Boltz run is made in a comma-separated value file with suffix ".bzlig" and you can open this file to show the table of results. But in cases where this file is missing you can recreate the table with this command which searches the prediction results extracting the scores, and searches the ".yaml" files to find the SMILES strings for the ligands.
> cd C:\Users\username\boltz\Scripts > pip.exe uninstall torch > pip.exe install torch --index-url https://download.pytorch.org/whl/cu118
$ cd ~/boltz2/bin $ ./pip uninstall torch $ ./pip install torch --index-url https://download.pytorch.org/whl/cu118