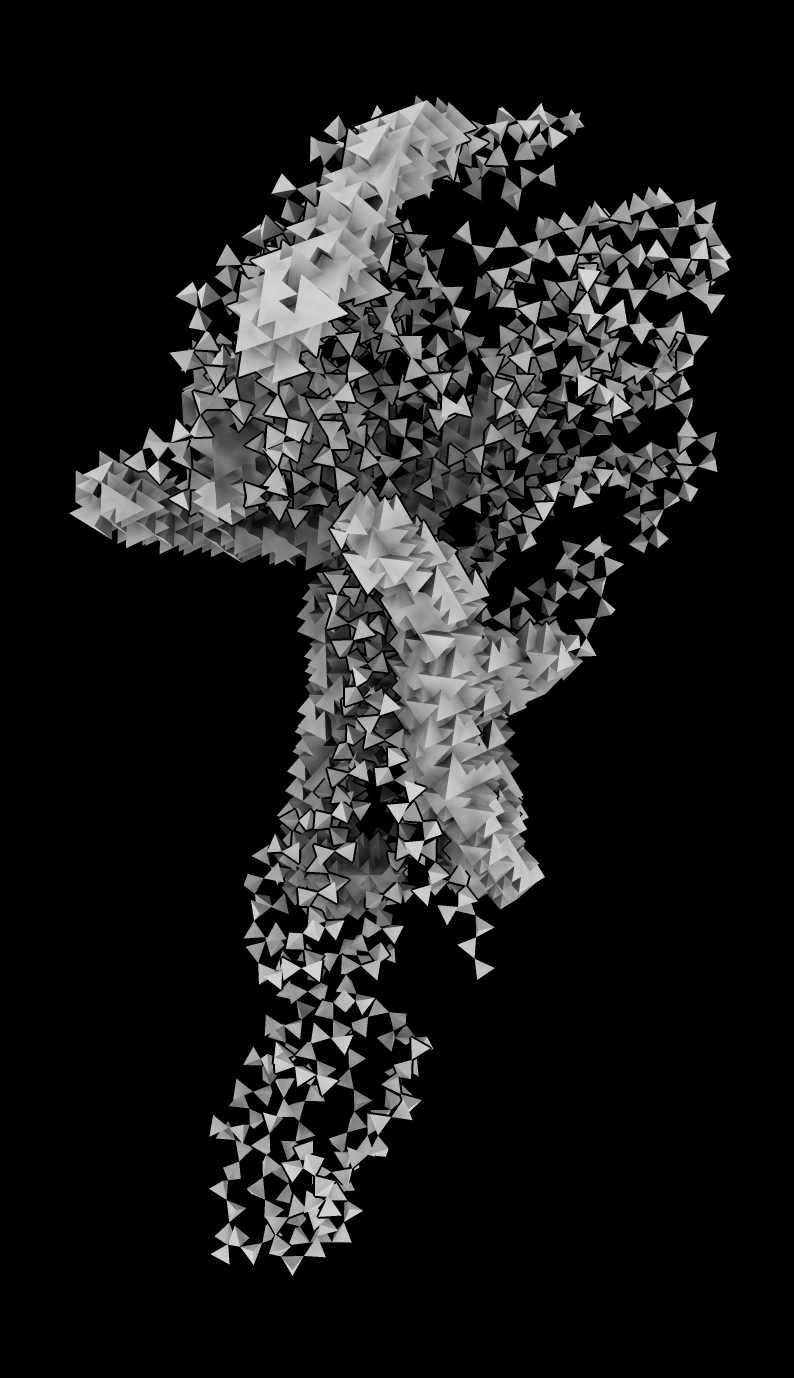Work in progress November 1, 2023 - January 17, 2024
Tom Goddard
January 18, 2024
Group meeting
Topics
- AlphaFold
- Collaborations
- Virtual Reality
- Miscellaneous
AlphaFold
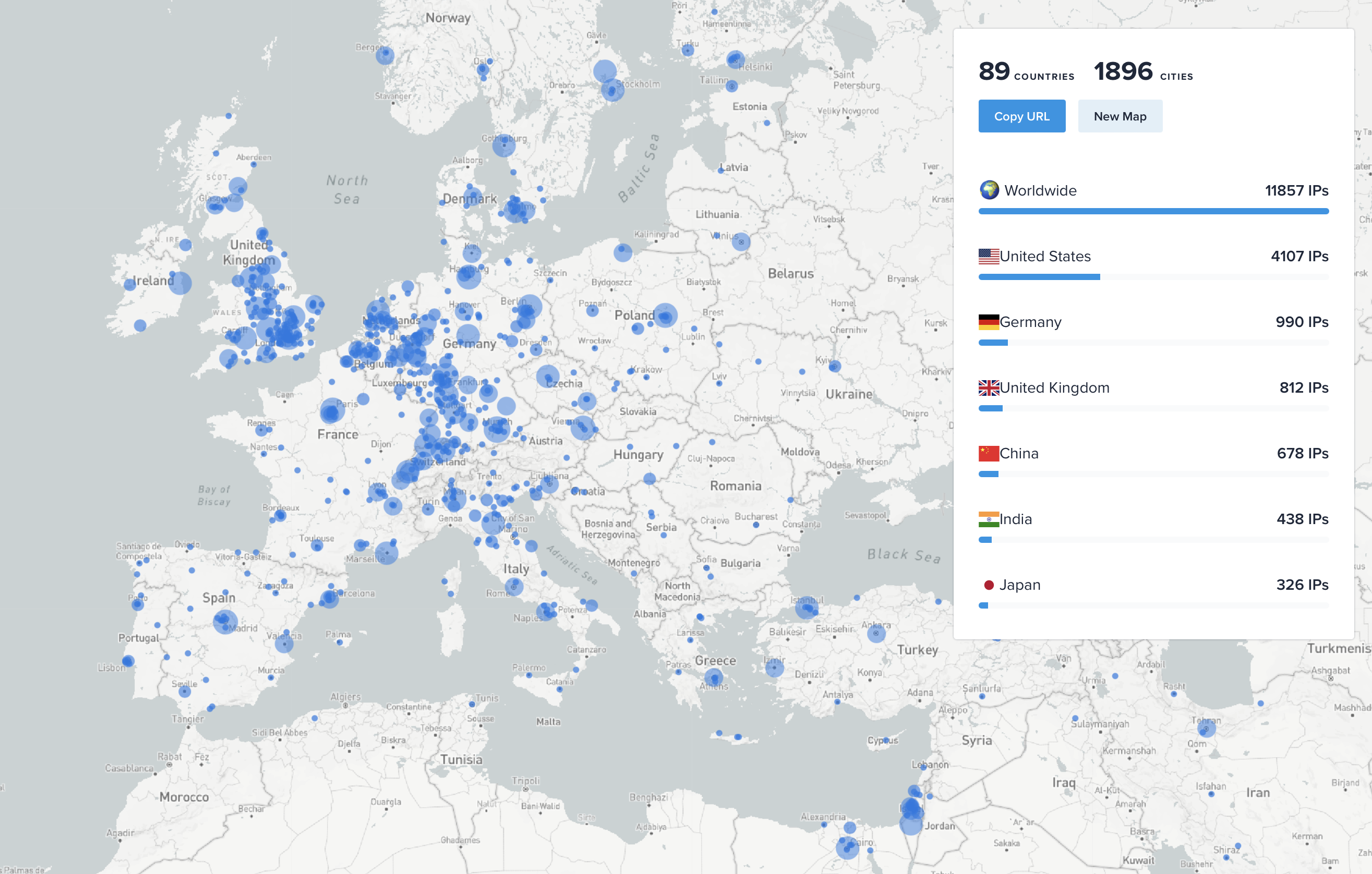
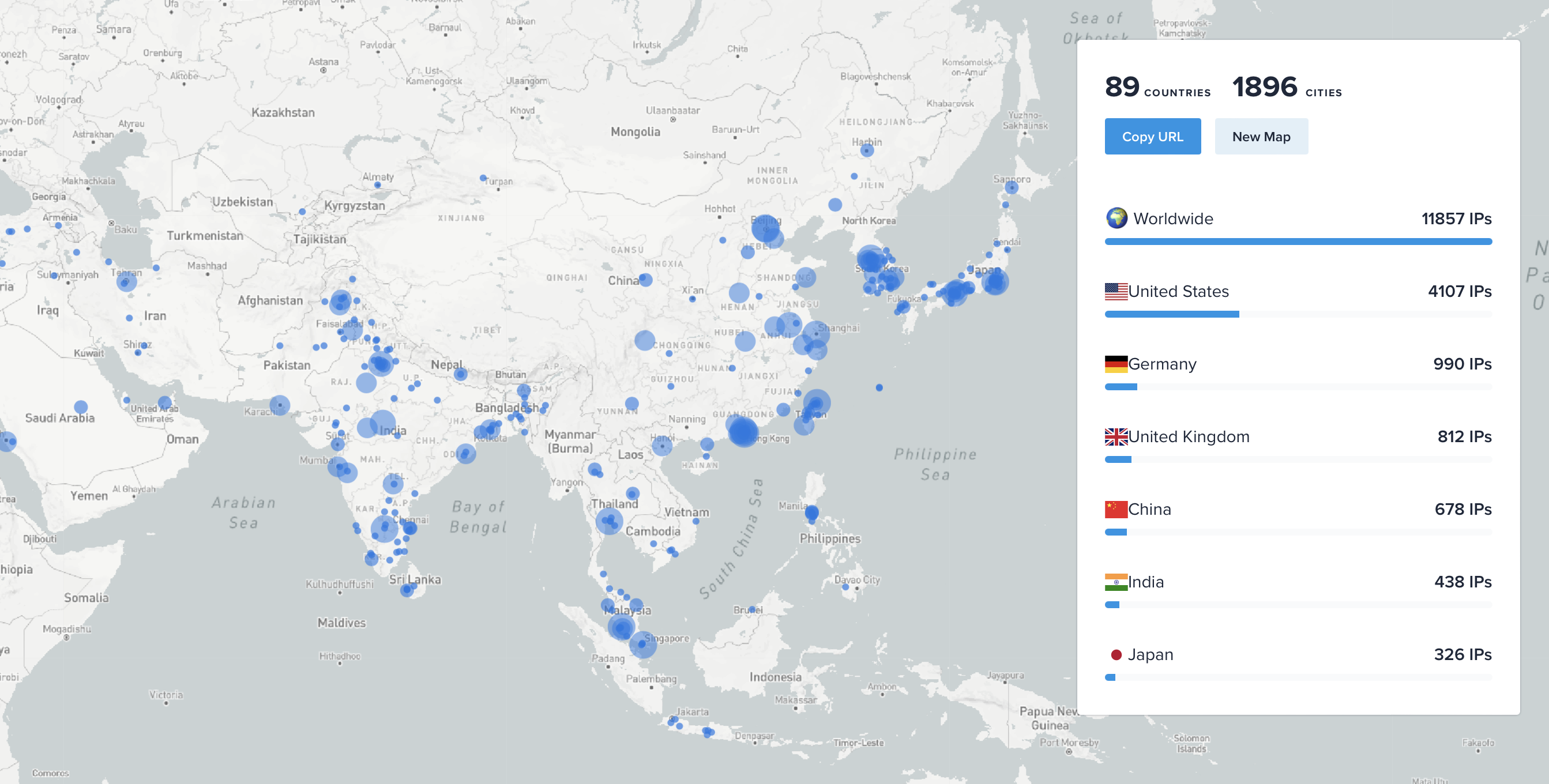
ChimeraX AlphaFold predictions on Google Colab
- 450 Alphafold predictions per day.
- 68,000 predictions made by 12,000 unique IP addresses in last 5 months, August - December 2023.
- This is would require about 10 GPUs in constant use assuming uniform usage throughout the day
and average prediction time at 30 minutes,
| ChimeraX Alphafold prediction IP addresses Aug-Dec 2023
|
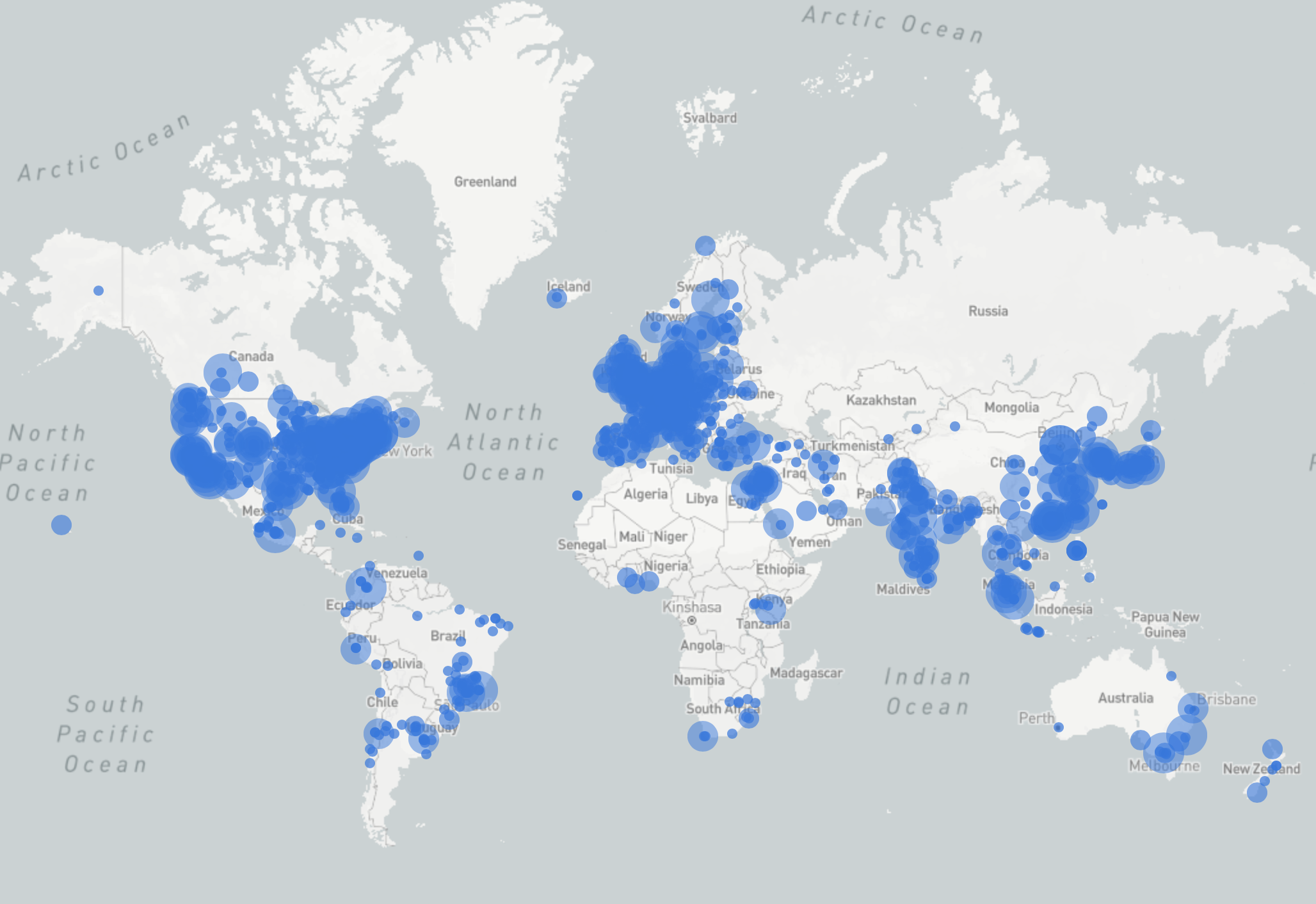
| 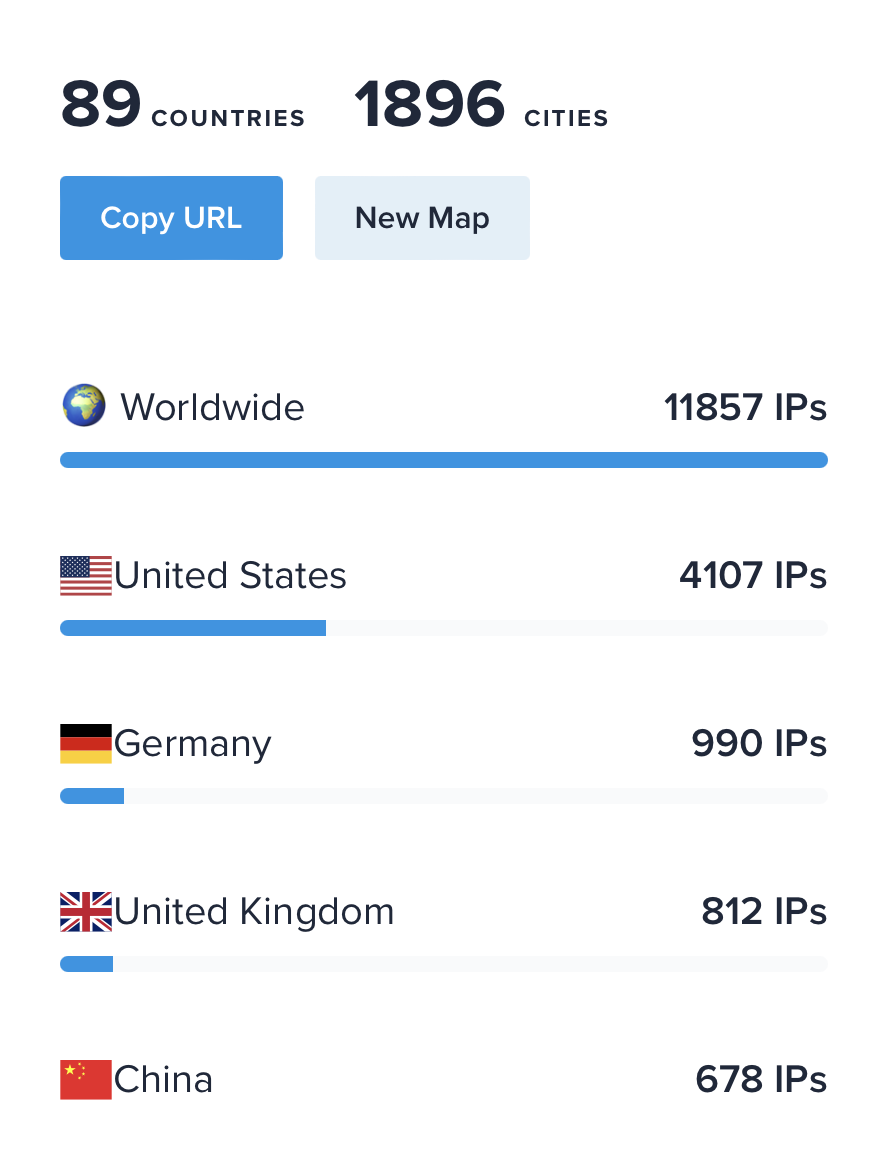
| 
|
Alphafold on Google Colab requires frequent maintenance
- ChimeraX Google Colab AlphaFold prediction service breaks every month or two.
- December 25, 2023: Failure caused by Biopython 1.82 update removing SCOPData class (bug 10402). Pinned Biopython to 1.81.
- November 8, 2023: Failure caused by a jaxlib error introduced in versions > 0.3.25 (bug 10115). Added jaxlib code patch with sed.
- November 6, 2023: Failure caused by Google Colab updating CUDA version from 11 to 12 requiring newer jaxlib for AlphaFold (bug 10094). Updated jaxlib to use Google Colab version (0.4.16).
- June 28, 2023: Failure caused by new hhsuite changing required Conda version breaking OpenMM (bug 9265). Made hhsuite install not update Conda.
- ...
- The breakage is caused by two factors:
- Google Colab updates their packages (e.g. CUDA) without any announcement.
- ColabFold and AlphaFold dependency versions are not pinned so updates break them.
Alternative AlphaFold prediction hardware is too slow
- CPU only.
- Mac ARM GPU, Metal.
- Wynton cluster.
Prediction on CPU without Nvidia GPU is extremely slow
- CPU only prediction using 5 cores is about 60 times slower than Nvidia 3090.
- 976 amino acid RIM23 homodimer.
- 1 minute 39 second on Nvidia 3090 GPU (quillian.cgl.ucsf.edu).
- 126 minutes on an Intel i9-9900KF (3.6 GHz, 8 cores, quillian.cgl.ucsf.edu, 64 GB)
- 104 minutes on an Intel i9-13900K (3 GHz, 24 cores, minsky.cgl.ucsf.edu, 64 GB memory)
Prediction on Mac M1 GPU
AlphaFold 8FF2 chain A prediction: left jax-metal, middle Mac M1 CPU, right PDB structure.

- Apple started a port of jax GPU library to Metal.
- PyPi package jax-metal.
- This should allow running AlphaFold on Mac GPUs.
- I tried jax-metal 0.0.4 and it ran without error on a short peptide (40 amino acids, PDB 8ff2_a).
- Produced completely wrong structure with all residues on top of each other.
- Running on Mac with CPU only produced a valid structure.
- Jax-metal prediction was slower than CPU only which was over 100 times slower than Nvidia 3090.
- jax-metal 0.0.5 released on Dec 21, 2023. Should test that.
ColabFold cannot run on Wynton
- ColabFold is 5 to 10 times faster than AlphaFold with equal quality.
- Wynton can't run ColabFold due to lack of internet access on compute nodes.
- ColabFold require internet access to run sequence alignments on massive cloud servers.
Predicting protein-protein interactions better
- To get more accurate dimer interfaces try 20 recycles according to 2022 Nature article.
- Normal AlphaFold "recycles" is 3.
- Tried on 21 dimers in RIM101 pathway of 6 proteins.
- Found 8 dimers, same as with 3 recycles, but one dimer added and one lost.
- Took 5 times longer, 69 hours, versus 14 hours with 3 recycles.
Predicting protein-protein interactions faster
- To make more dimer predictions use only 1 recycle (twice as fast) and predict only 1 of 5 models.
- Tried on 21 dimers in RIM101 pathway of 6 proteins.
- Found 4 dimers, half as many as with 3 recycles.
- Took 1.5 hours, 10 times faster than with 3 recycles.
Collaborators interested in AlphaFold for finding protein-protein interactions
- Hiten Madhani (UCSF), writing R01 grant including AlphaFold on RIM101 pathway.
- John Gross lab at UCSF. I will present at their group meeting Feb 6, 2024.
- Zhen Chen setting up new lab at Caltech.
- Dengke Ma (UCSF) wants to search C elegans all AlphaFold predictions for membrane cysteines involved in electron transport.
- David Fay, U Wyoming, NIH INBRE center which data science core. Asked for AlphaFold workshop.
- Klim Verba (UCSF), interested in kinase interactions.
- Bob Stroud (UCSF), tuberculosis transmembrane protein dimerization study.
- I am working on a ChimeraX "alphafold dimers" command to setup runs using local ColabFold.
- Also developing a "alphafold interactions" command to analyze interfaces.
Collaboration and Outreach
NMR restraints
| 
Segmented light microscopy
| 
Christmas card
| 
Tetrahedron proteins
| 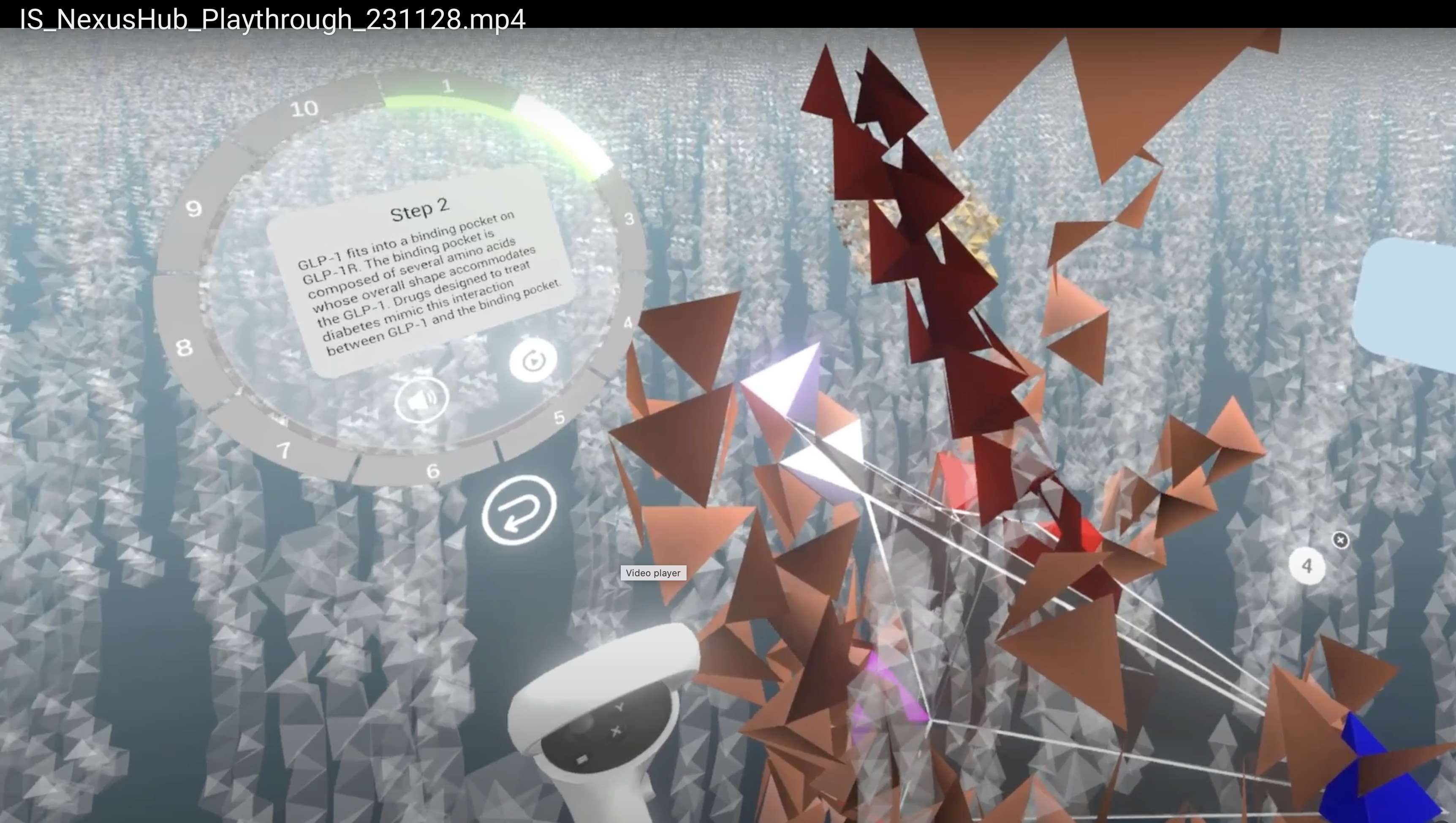
Inner Space VR
|
- Kristen Browne, Added vrml export for NIH 3D.
- Aashish Manglik, John Gross. Gave Macromolecular Methods class new 2-hour Chimerax tutorial.
Added ability to fetch/show NMR constraints from PDB
- Randy Read wrote emplace_local manuscript. Reviewed.
Compared to Chimerax fit search capability, how many positions to search.
- Helen Berman, Bryan Zhang.
Tested Inner Space VR experience and discussed in a dozen emails with Bryan.
- Roshan Kumar, Jitin Singla. Helping make Chimerax command for tetrahedron residue depictions.
- Brinda Vallat. Reviewed IHM PDBDev manuscript.
- Sam Lord. Segment fluorescently labeled proteins in archael cells in 3D light microscopy.
Made Chimerax Python script
using Segger segmentation.
- Tug interactive molecular dynamics uses
- Haleigh Miller (DeRisi lab).
Pull AlphaFold antibody model toward known structure to see if engineered loops long enough
- Thomas Marlovits, build cyclic peptides, drag ends together to get reasonable bond lengths.
- Added OpenMM compute platform option (cuda, opencl, cpu) for faster MD.
- Oliver Clarke visited November 2, intesting Ankyrin talk, many feature requests.
- Made christmas card, human myosin thick filaments.

Quest 3 docking station
|
Virtual reality
Got two Quest 3 headsets
- Expected to be faster than Quest 2, but cannot handle scenes with more triangles in LookSee.
- Docking station for wireless charging including hand controllers is an improvement.
- Got elite headstrap, silicone face pads for easy cleaning.
- Each headset has its own Meta account.
- Next step is to run VR demos in the Genentech Hall attrium showing researchers their data.
Looksee improvements
- Added two new features suggested by Phil Cruz.
- Option to keep models aligned, works with meetings too.
- Play model series by tilting thumbstick.
- Distribution via Meta AppLab has hit a snag.
- Requires organization verification by drivers license, Meta won't recognize academic organizations.
- Currently can only make pre-releases on AppLab.
- 24 users have joined prerelease channel (including UCSF and NIAID).
NIAID VR medical image demo
- Helped Phil Cruz improve VR user interface for Radiology Society of North America demo.
- Made windowing update volume histogram instantly.
- Use buttonpanel to create custom buttons to press in VR that run commands.
- Use tilted slab volume rendering, most effective for visualizing medical images.
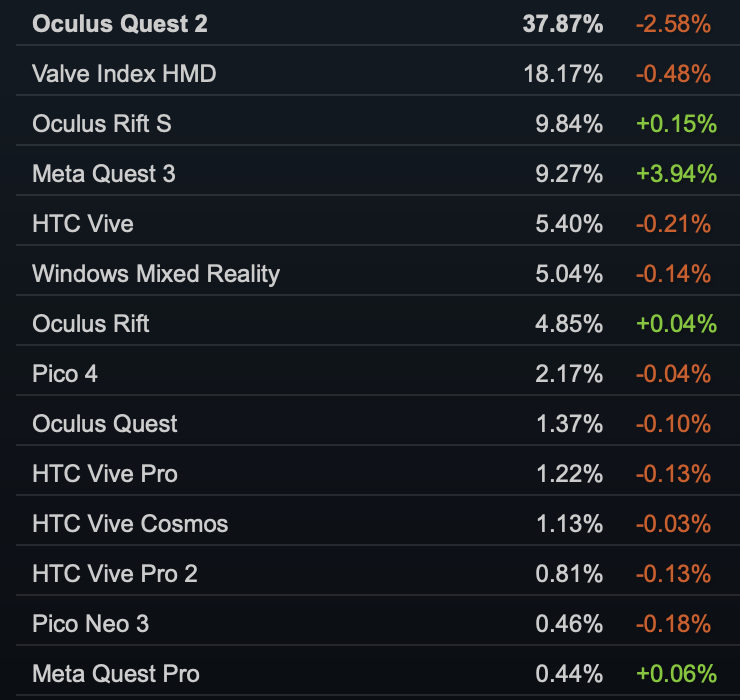
Steam VR headset share December 2023
|
Standalone VR replacing PC VR
- Apple Vision Pro releasing February 2, 2024, $3500.
- Microsoft deprecated Windows Mixed Reality in December 2023, will be removed in 2026.
- Current generation PC VR headsets are old (Valve Index 2019) or have been discontinued (Oculus Rift S).
- Most sentiment online is that PC VR is dead, replaced by Meta Quest standalone.
- Although Meta Quest can work with PC VR, gamers complain there is no new software taking advantage
of high power PC graphics since only profitable market is standalone (mobile graphics).
Miscellaneous
- ChimeraX 1.7 released
- New Mac Studio desktop. M2 Ultra, 24 CPU cores, 60 GPU cores.