Progress report May 11, 2023 - July 5, 2023
Tom Goddard
July 6, 2023
Group meeting
Possible collaborative projects
Dyche Mullins and Laura Cheradame - segmenting filaments in tomograms
- Met with Dyche and Laura and looked at tomograms DNA damage in mouse neurons.
- They are interested in trying machine learning segmentation to distinguish cells
with damaged DNA from normal cells.
- By eye we can see filaments in the damaged DNA puncta.
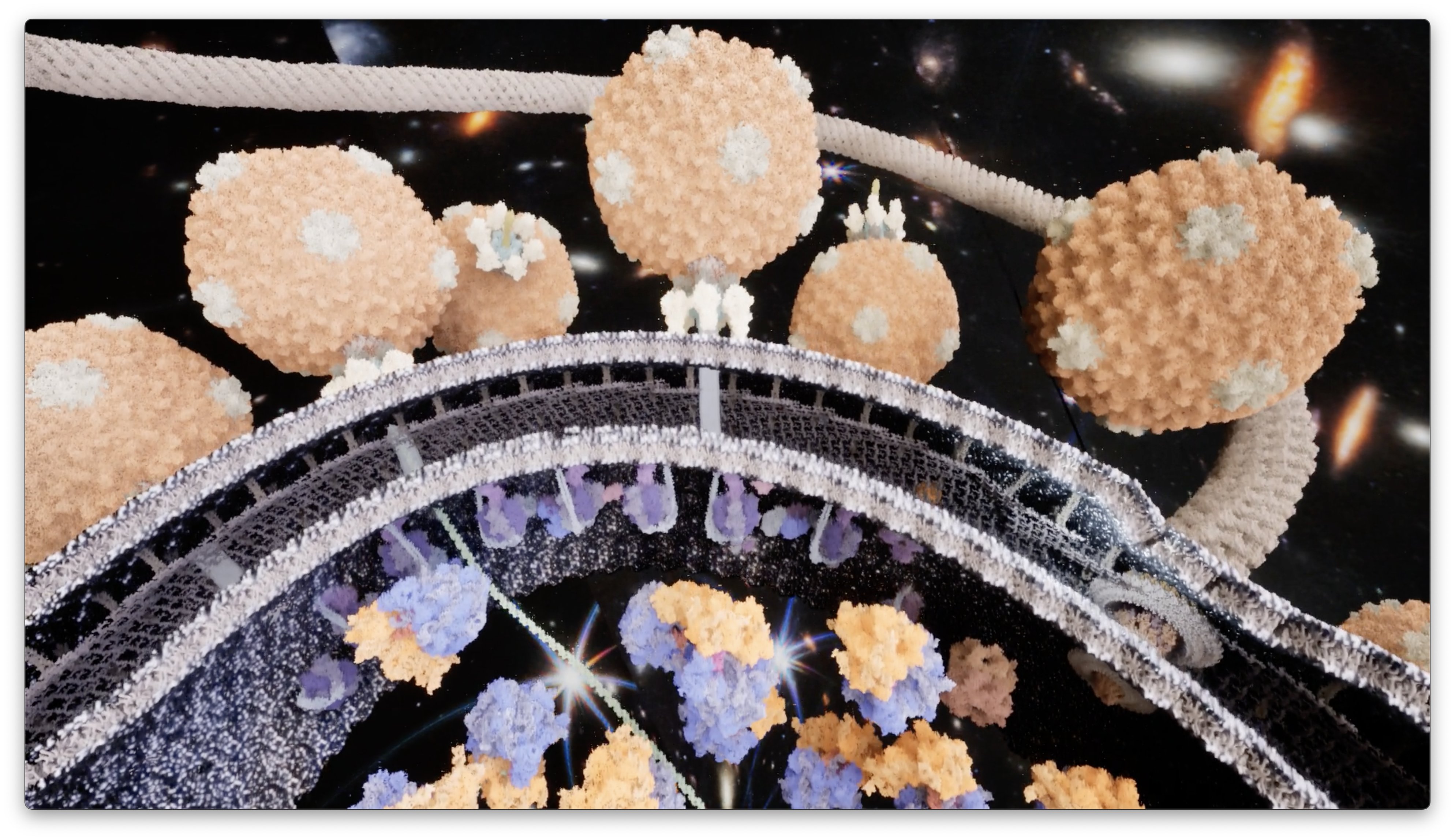
Muyuan Chen in Wah Chiu's lab, virus animations
- Muyuan used ChimeraX exported models with Unreal Engine level-of-detail capabilities
to animate virus scenes with tens of millions of atoms. Video.
Helen Berman and Jitin Singla - tetrahedron protein depictions
- Meeting Helen by Zoom about once every few months for her Inner Space project at USC, animating pancreatic beta cells for VR.
- Jitin Singla's student writing ChimeraX bundle to depict each residue as a tetrahedron.
- Next Zoom meeting July 18.

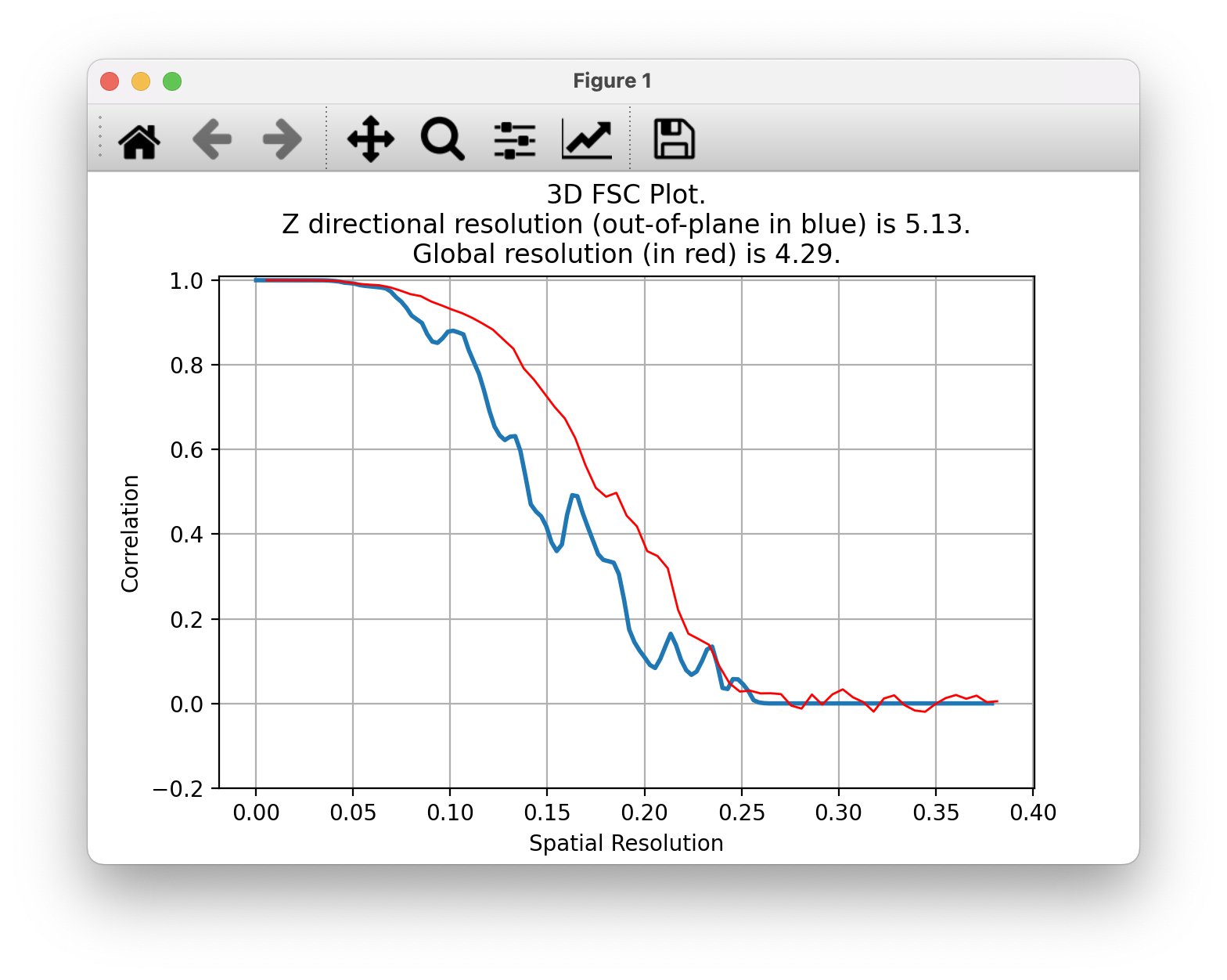
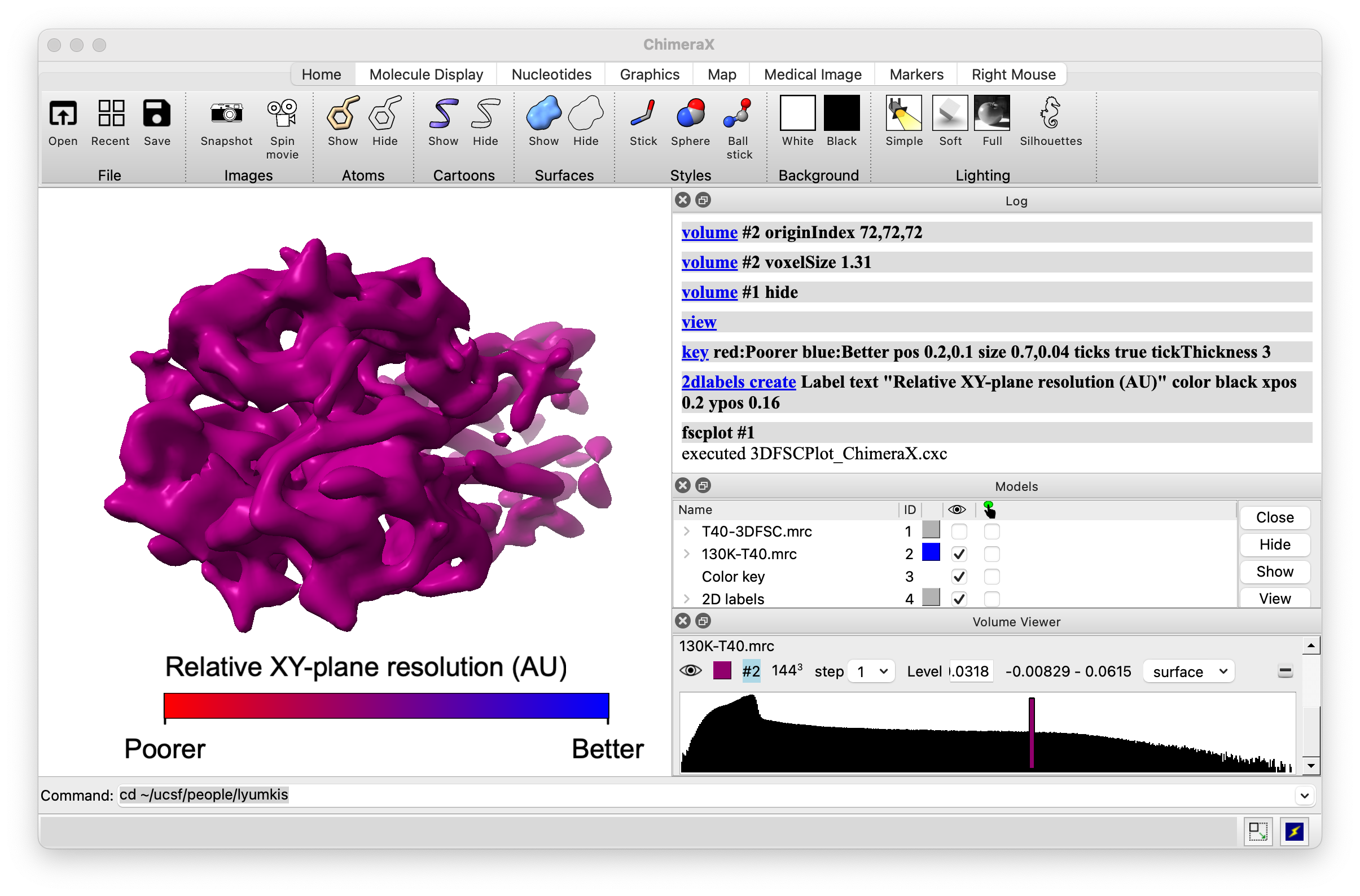
Dmitry Lyumkis, Salk Institue cryoEM, fscplot extension
- I helped update Chimera fscplot tool to ChimeraX.
Max Krummel, embed 3D light microscopy in web pages
- Made web page explaining how
to embed ChimeraX GLTF scene using Google Model Viewer.
- Max also interested in showing 3D tumor cells with Quest VR.
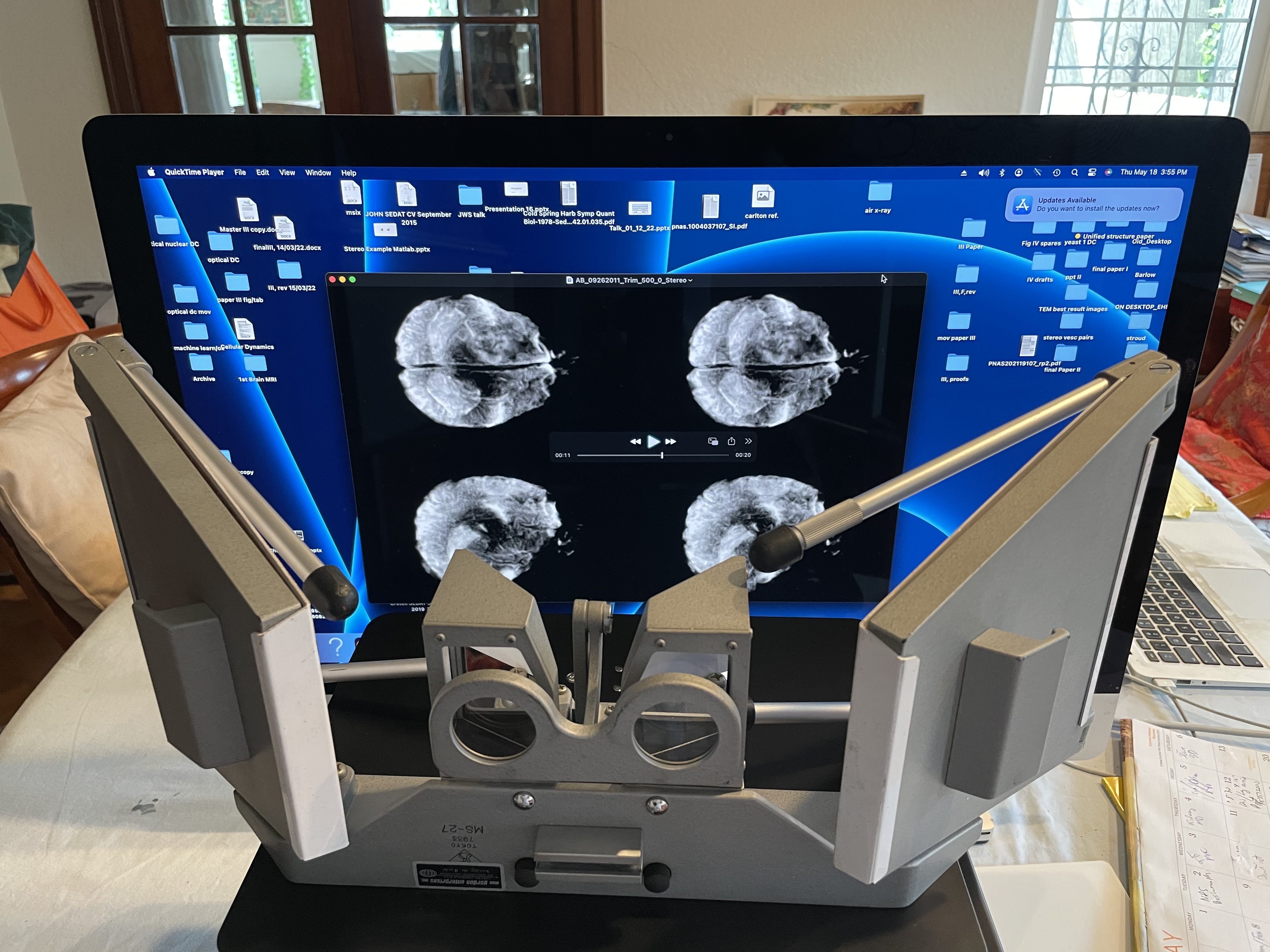
John Sedat, stereo visualization of low dose CT scans
- John has noise filtering methods to allow medical CT scans at 10x lower dose.
- Trying to promote stereo visualization in clinical setting.
- Met with John and had a few phone calls to discuss use of VR for stereo viewing.
Tristan Croll, ISOLDE on Apple Vision Pro AR headset
- Discussed visionOS for running ISOLDE.
- Tristan says Altos Labs software head interested and could hire a couple Apple developers.
- Discussed continuous Chimerax scene updates to standalone VR to avoid speed bottlenecks.

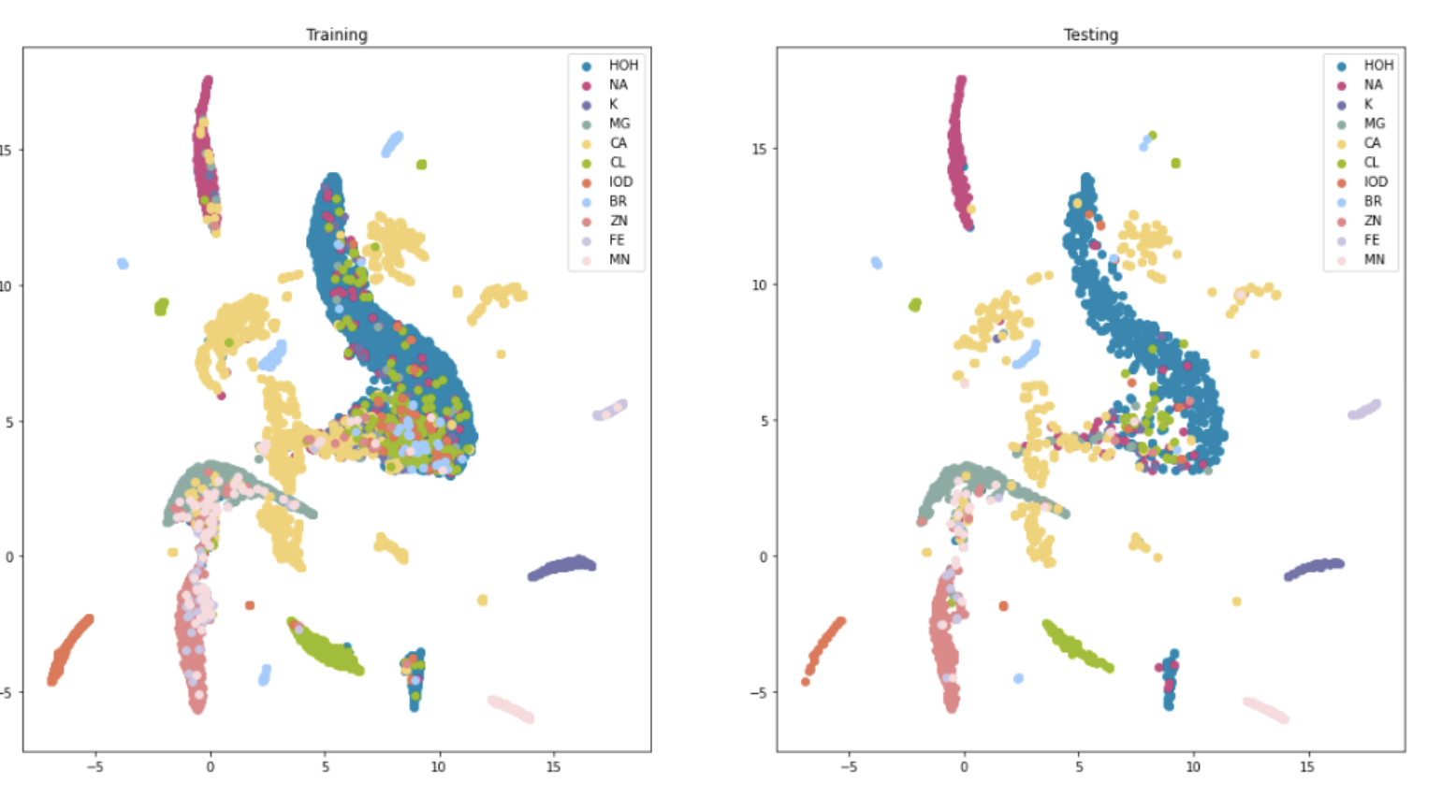
Selina Liu, locating ions in cryoEM maps
- Selina developed ML in Keiser lab to identify ions in cryoEM maps near atomic model.
- This might make an interesting ChimeraX tool.
- Similar to Phenix douse that finds waters in cryoEM maps near atomic model.
- Use UMAP visualization of training and testing results.
NIAID NIH 3D web site and GLTF and surface resolution
- Tried NIH 3d web site, downloaded cellpack virus and viewed in ChimeraX.
- Made various ChimeraX GLTF export improvements for Kristen Browne.
- Remove vertex colors for single color objects.
- Added flat_lighting option.
- Added backface_culling option.
- Investigated jagged NIH 3D molecular surfaces in meetings and emails.
- Sped up million atom molecular surface calculation 100-fold.
- Considering mesh decimation, sharp-edges, offering multiple NIH 3D resolutions.
- Investigated GLTF vs ChimeraX color differences, linear RGB vs sRGB, Kristen Browne
 | 
|
| GLTF viewer colors | ChimeraX colors
|
AlphaFold updates
- Colab now offers A100 GPUs at market rate ~$1-2/hour that ChimeraX AlphaFold prediction can use.
- Tested running 2000 residue alphafold prediction with A100 gpu on google colab.
- Tweeted
and made YouTube video showing how to use A100.
- Added maxPae option to alphafold contacts command, Ah-Ram Kim
Alphafold prediction web service maintenance
- Google Colab updates break AlphaFold prediction several times per year.
- Colab update from Python 3.9 to 3.10 broke energy minimization because old OpenMM required
by AlphaFold was not available on 3.10. Required ColabFold fixes to use newer OpenMM.
- Reported new ColabFold bugs to Sergei Ovchinnikov reports, logging.
- Sergei accepted my earlier ColabFold pull request allowing me to simplify Chimerax prediction script.
- Fixed alphafold prediction openmm install error cause by bioconda update.
- Helped a few users recover failed alphafold downloads.
Meetings
Stanford cryoEM Workshop
- Gave 2 hour intro cryoEM tutorial to 50 in-person and 50 online participants, atp synthase,
at SLAC cryoEM workshop hosted by Wah Chiu.
- 30 minutes of questions and requests at SLAC workshop.
- Ported mcopy command from chimera to chimerax
- Ported volume filter gui, Mike Schmid request
- Added volume sharpen command.
Fraser lab / Phenix / ChimeraX 1 day ensemble modeling meeting
- Progress in software for ensemble modeling.
- In maps with better than 1.8A resolution can see alternative atom locations for 40% of residues.
How do we improve the modeling of ensemble-based models?
Location: UCSF Mission Bay Genentech Hall S202
9:00 am: Introduction (Jaime Fraser & Paul Adams)
9:15 am: Multiconformer modeling of X-ray and cryoEM structures with qFit (Ashraya)
10:15 am: Break
10:40 am: Measuring map-model fit and resolvability with Q-scores (Greg Pintilie)
11:10 am: Phenix Team: Quantum Refinement (Nigel)
11:45 pm: Discussion on new CIF model with alt confs (Jaime)
12:10 pm: Lunch at SF Kebab
1:10 pm: ChimeraX: Ensemble capabilities (Eric Pettersen)
1:40 pm: Multi-part bulk solvent and Model-to-Map Fit Validation (cryo-EM) (Pavel)
2:10 pm: Modeling partially occupancy water molecules (Stephanie)
2:40pm: Bayesian optimization of multiple models in cryoEM maps (Matt Hancock, Sali lab)
3:10 pm: Closing remarks
- Tried Quest VR visualization of large number of alt-locs (33%) in transaldolase PDB 6zx4.
- Standalone VR may be useful for discussions of alt-loc functions.

Quest 2 multi-person AR meetings
- Added multiperson AR or VR meetings to Quest molecular viewer app LookSee.
- 2 or more participants see the same molecules, maps and can move and scale.
- Any participant can open new models which are copied in seconds to others.
- Mike Schmid at Stanford will try it with tomograms.
- Andreas Bueckle, NIAID VR user study designer will try it.
- Learned about async/await programming (C#, also in Python) to handle network meeting messages.
LookSee user interface for starting and joining a meeting:
Miscellaneous ChimeraX improvements
- New ChimeraX recipe how to realign map symmetry axis to z, for Tianyang Liu.
- Added move-to-center mouse mode, for Yang Lee.
- Color large structures when opened by polymer instead of single color.

- Made C-alpha RMSD coloring how-to video and tweeted, for Matthias Vorlander.
ChimeraX daily now using Python 3.11
- Zach updated ChimeraX from Python 3.9 to 3.11.
- Had to fix a handful of problems (e.g. no implicit casting float to int).
New computers
- New VR machine with Nvidia 3070, setup in corner in vizvault -- multi-person VR is working. Windows 11.
- New ML machine in treasure room, Nvidia 4090 (24 GB memory), Ubuntu 22. AlphaFold. Selina is training lung tumor ML on it.