NIAID contract progress October 6 - November 17, 2022
Tom Goddard
November 17, 2022
Group meeting
- NIAID contract progress
- Eric - NIH 3D pipeline
- Greg - small molecule CIF
- Zach - DICOM
- Elaine - documentation for contract features
- Tom - machine learning structure prediction for cryoEM
- CZI Imaging Institute electron tomography workshop
- ESMFold protein structure prediction from Meta
- AlphaFold prediction service for longer sequences
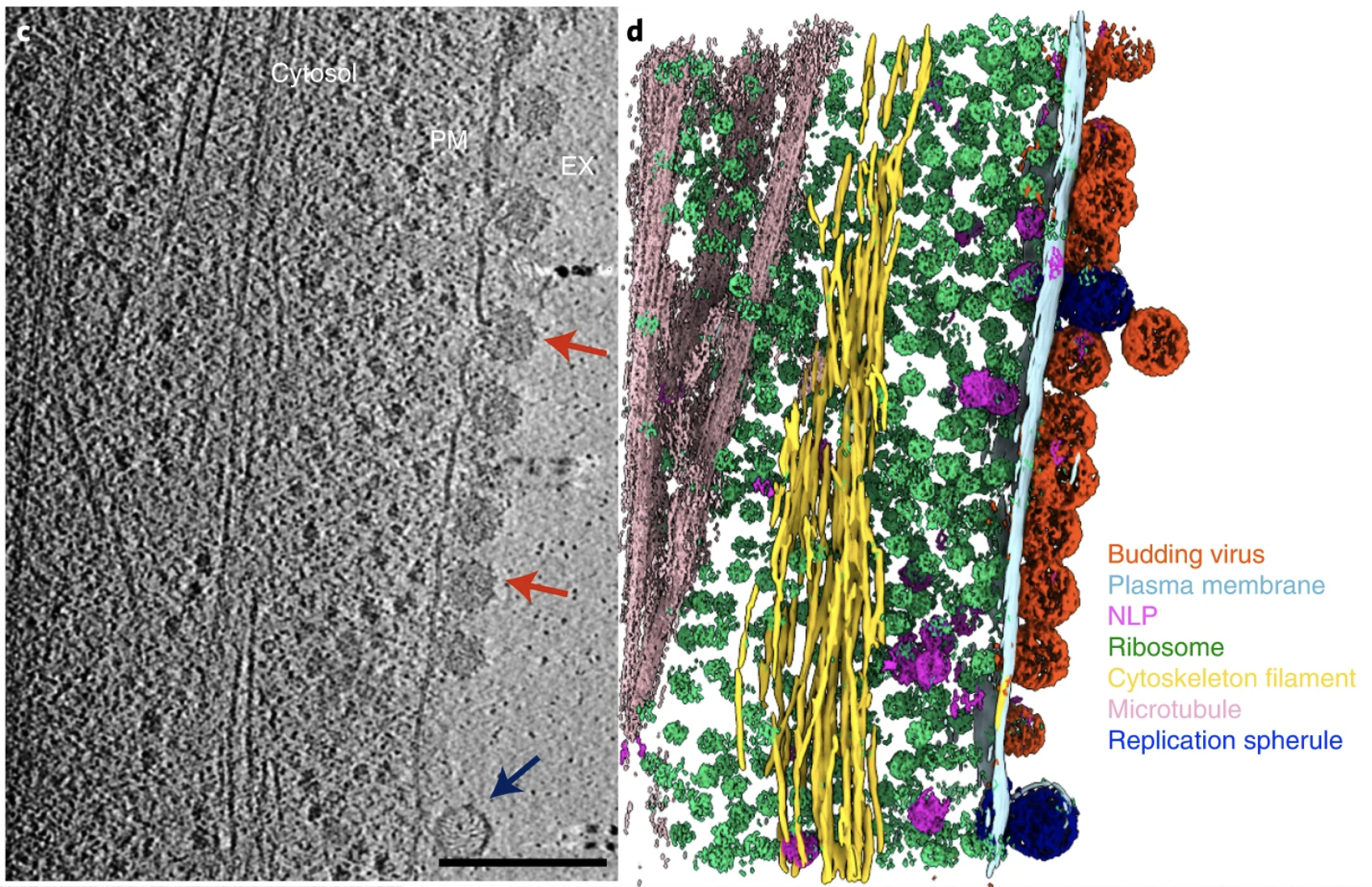
CZI Imaging Institute electron tomography workshop
| 
|
- Workshop to discuss what the new CZI Imaging Institute should do.
- About 50 participants, held at Mountain View 5-star hotel.
- Organized by Dave Agard, Bridget Carragher and Clint Potter, directors of the institute.
- Agenda and participants
- Lots of interesting participants, got to talk with 20 of them.
- Utz Ermel - ChimeraX ArtiaX developer, Goethe Univ Frankfurt
- Ellen Zhong - cryoDRGN developer, Princeton
- Alister Burt - Napari plugin developer, MRC-LMB
- Nick Sofroniew - Napari project leader, CZI
- Michael Schmid - ChimeraX VR expert with Wah Chiu, Stanford
- Wah Chiu - director of SLAC cryoEM
- Many others...
- Lots of machine learning methods presented or discussed: cryoDRGN, cell segmentation, IsoNet missing wedge filling.
- My talk
Visualizing Tomograms
focused on need for database of cell segmentations.
- Napari
- Napari has about 250 plugins for 2D and 3D microscopy image analysis.
- CZI gave $2 million in grants to develop plugins.
- Two Napari talks made strong pitch to develop future cryoET visualization using Napari plugins.
- Napari distributed as PyPi and Conda packages for developers and desktop app for users.
ESMFold protein structure prediction from Meta
- Meta released 600 million protein structure predictions, called ESM Metagenomic Atlas, on November 1, 2022.
- ESM stands for "Evolutionary Scale Modeling".
- Uses machine learning "language model" to predict structures 10 times faster than Alphafold, but less accurate.
- Does not use deep multiple sequence alignment as input, just single sequence.
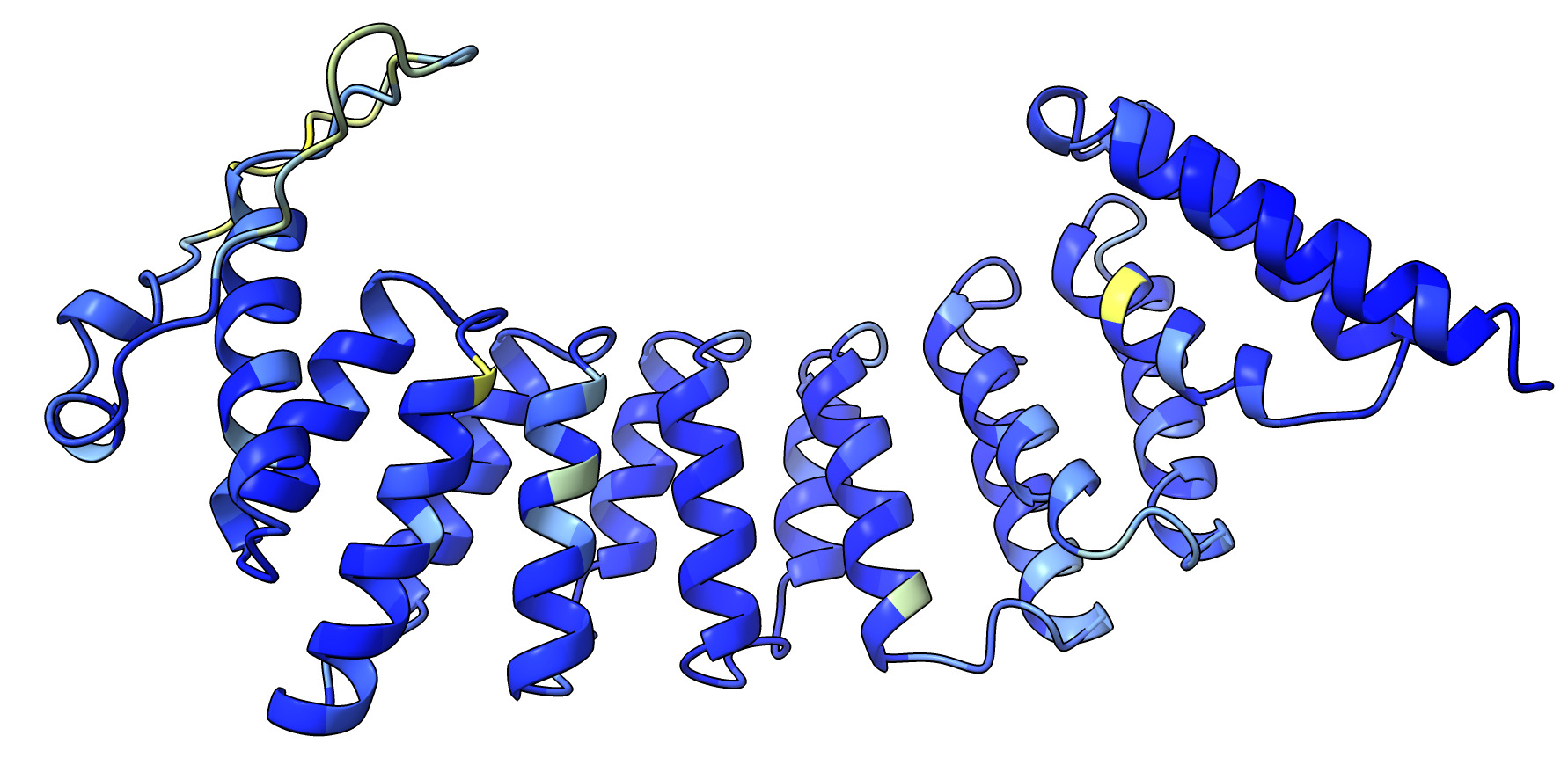
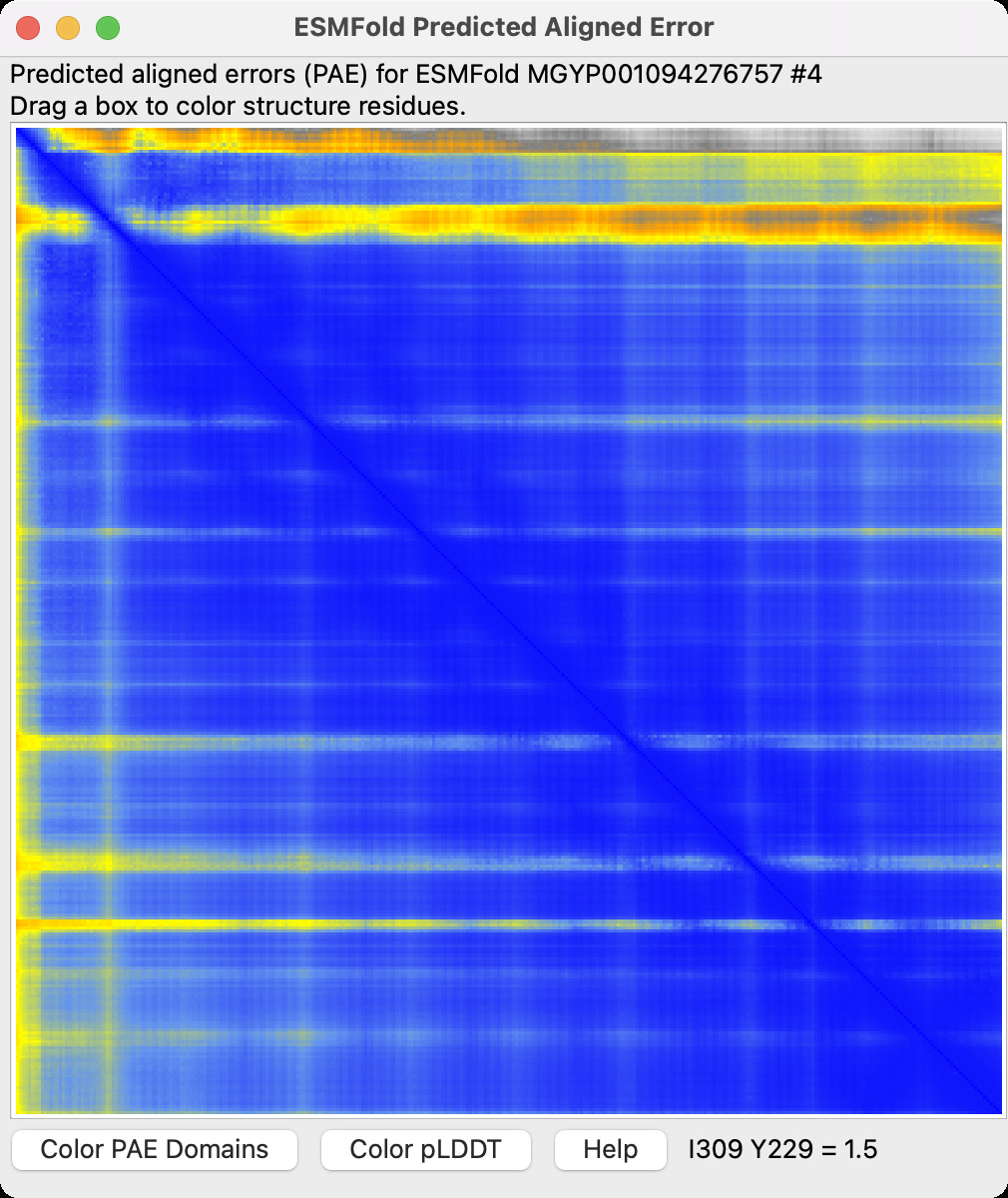
- Added ChimeraX command "esmfold predict" to predict structure from sequence
using Meta's server.
Example.
- Prediction server limits sequence length to 400 amino acids.
- Prediction made in under 30 seconds.
- Working on ChimeraX web service for Atlas sequence search (fast kmer search and blast).
AlphaFold prediction service for longer sequences

- Can we use new lab computer with 4 Nvidia A40 GPUs for ChimeraX AlphaFold prediction?
- Main advantage over Google Colab is ability to predict sequences longer than 1000, up to ~4000, using more GPU memory than available on Colab.
- Problem is long sequences are slow to predict, e.g. 28 hours for 3200 sequence length on Wynton.
- With only 4 GPUs few long sequence jobs (5-10) could run per day.
- Instead I think we should allow ChimeraX predictions to run on paid cloud GPU services: AWS, Lambda Cloud, Microsoft Azure, Google Cloud, ... many others.
- Researchers would have ChimeraX use their GPU virtual machine. Cost ~$2-3 / hour.