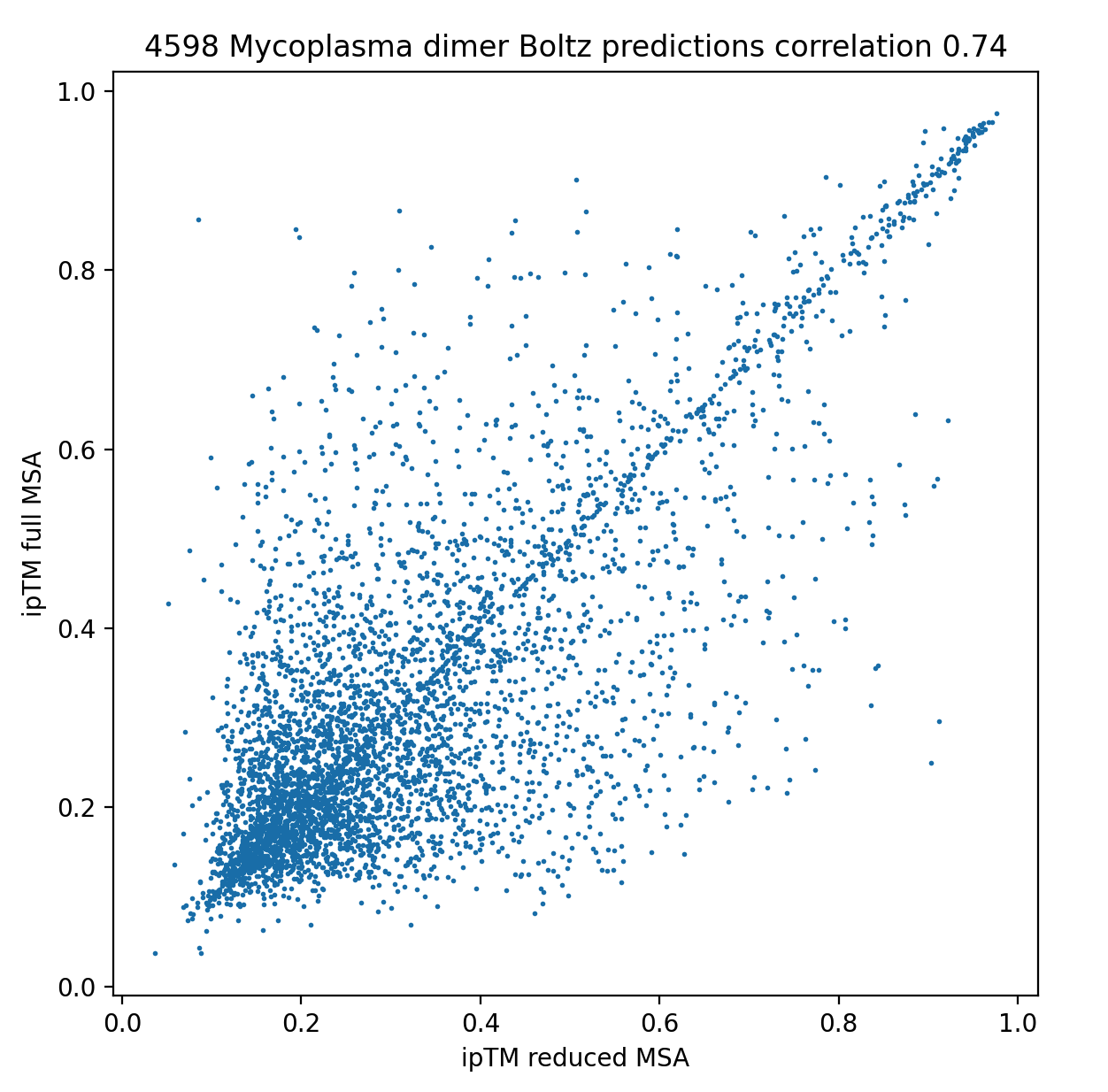
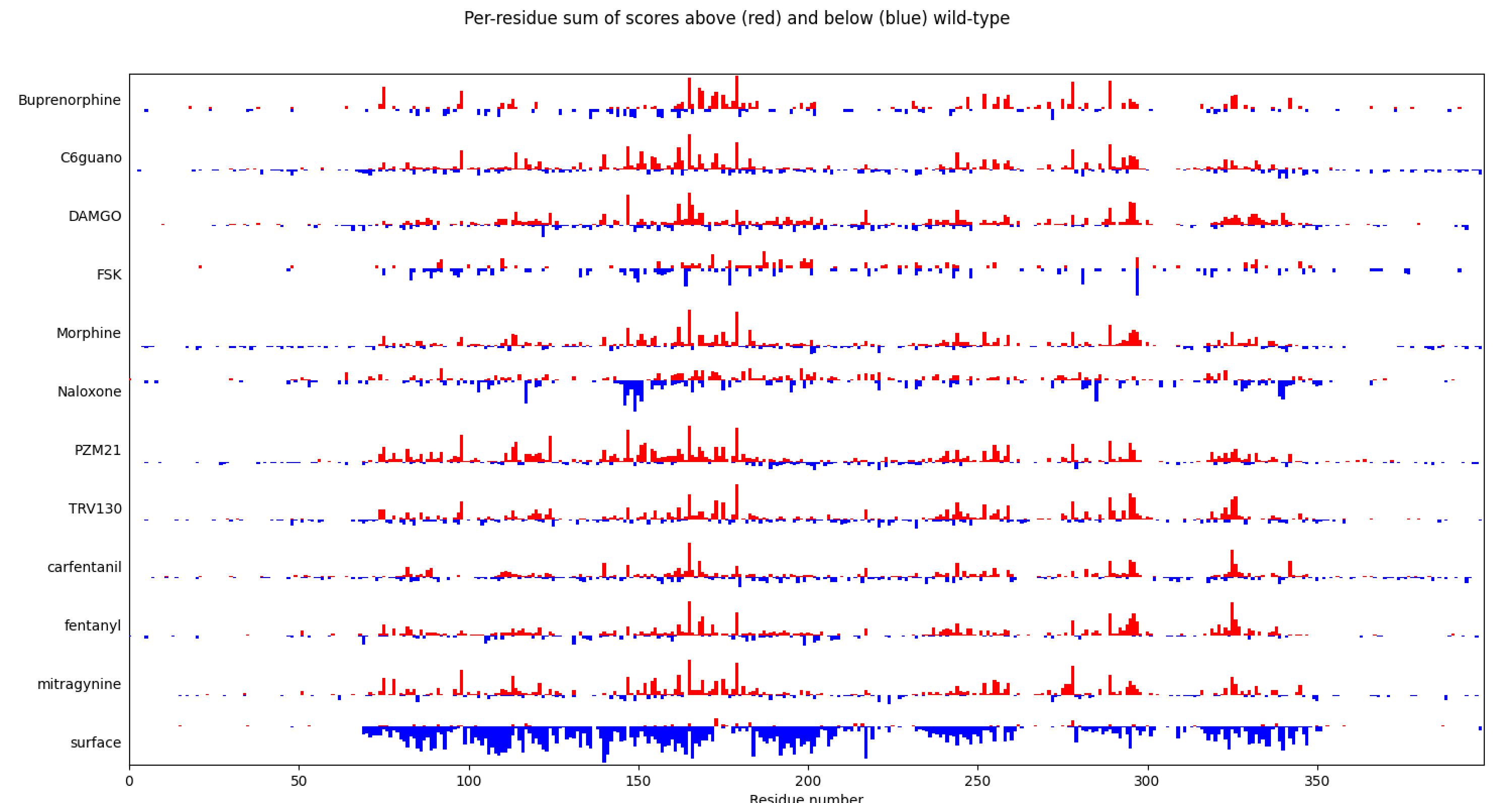
Correlation of ipTM interface quality scores
for 2 Boltz runs with reduced or full length MSAs.
Tom Goddard
October 23, 2025
Group meeting

Correlation of ipTM interface quality scores for 2 Boltz runs with reduced or full length MSAs. |
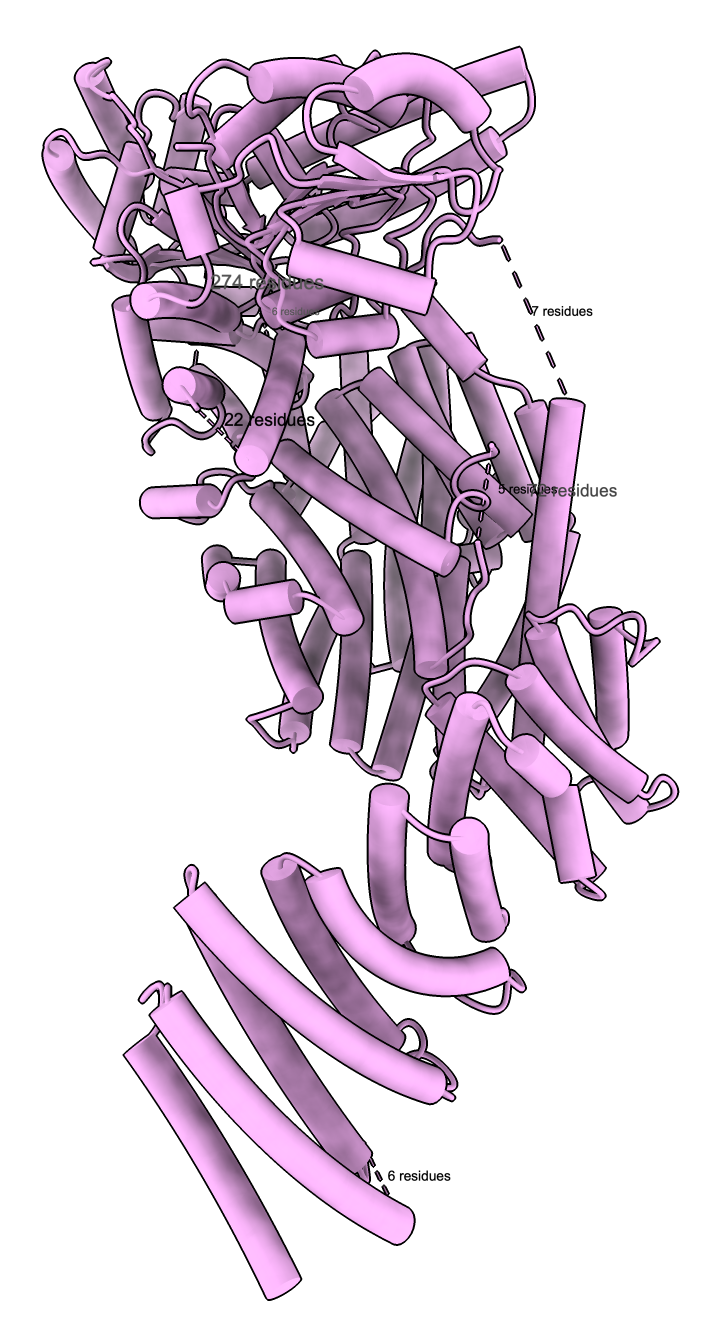
human separase PDB 9hma (cryoEM) 2200 amino acids | 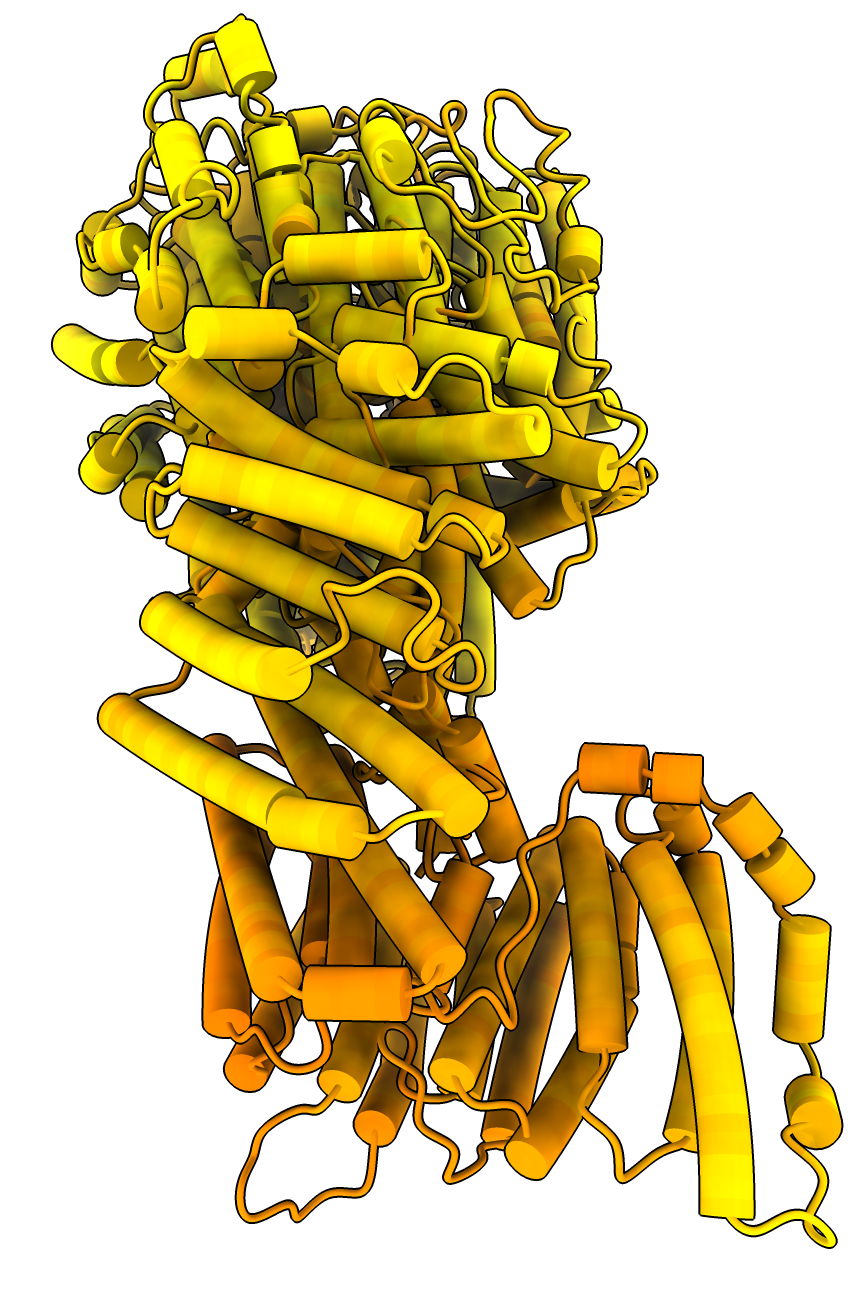
SimpleFold prediction 3B model wrong fold unphysical clashing helices | 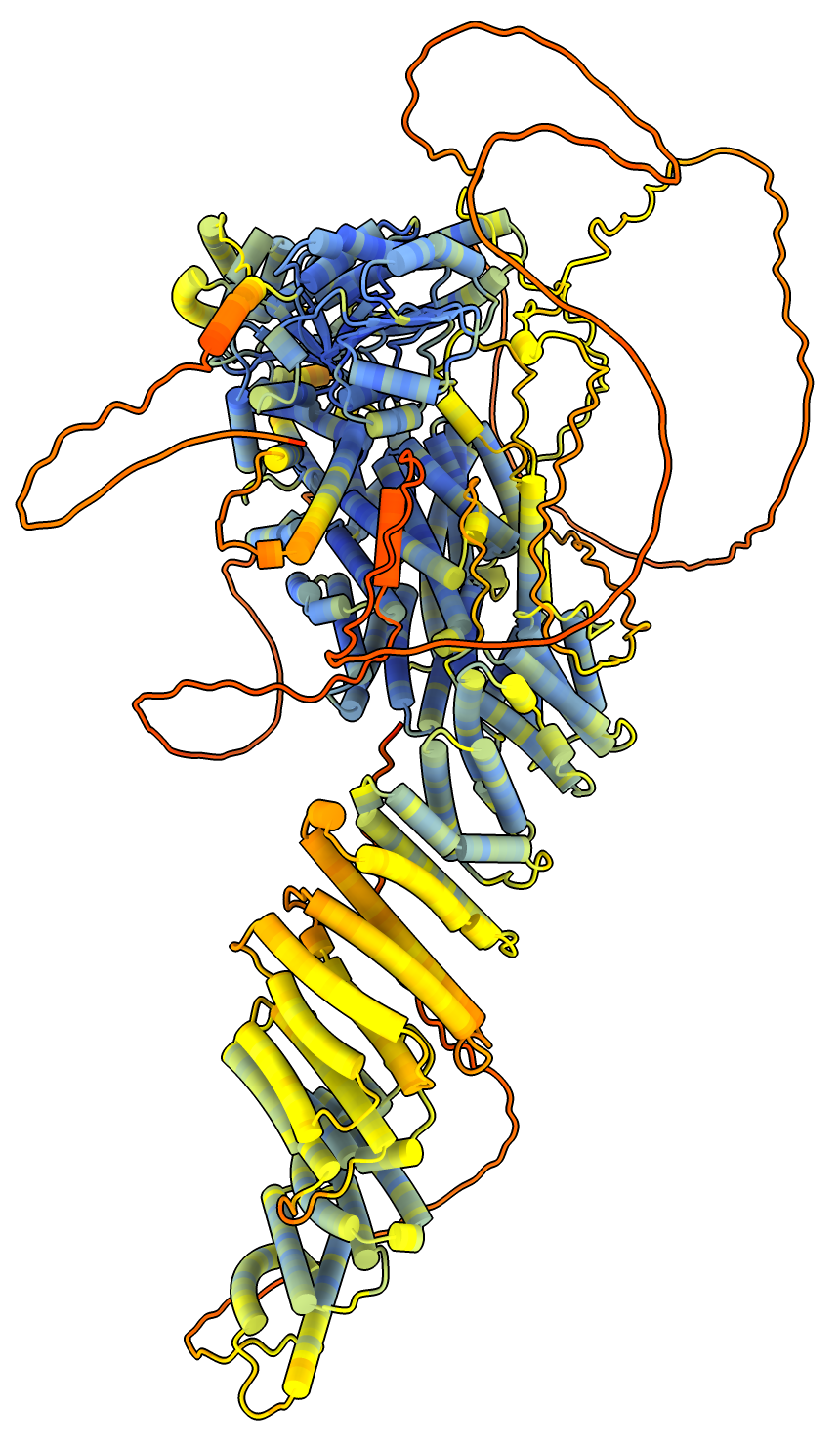
AlphaFold 3 prediction mostly correct |
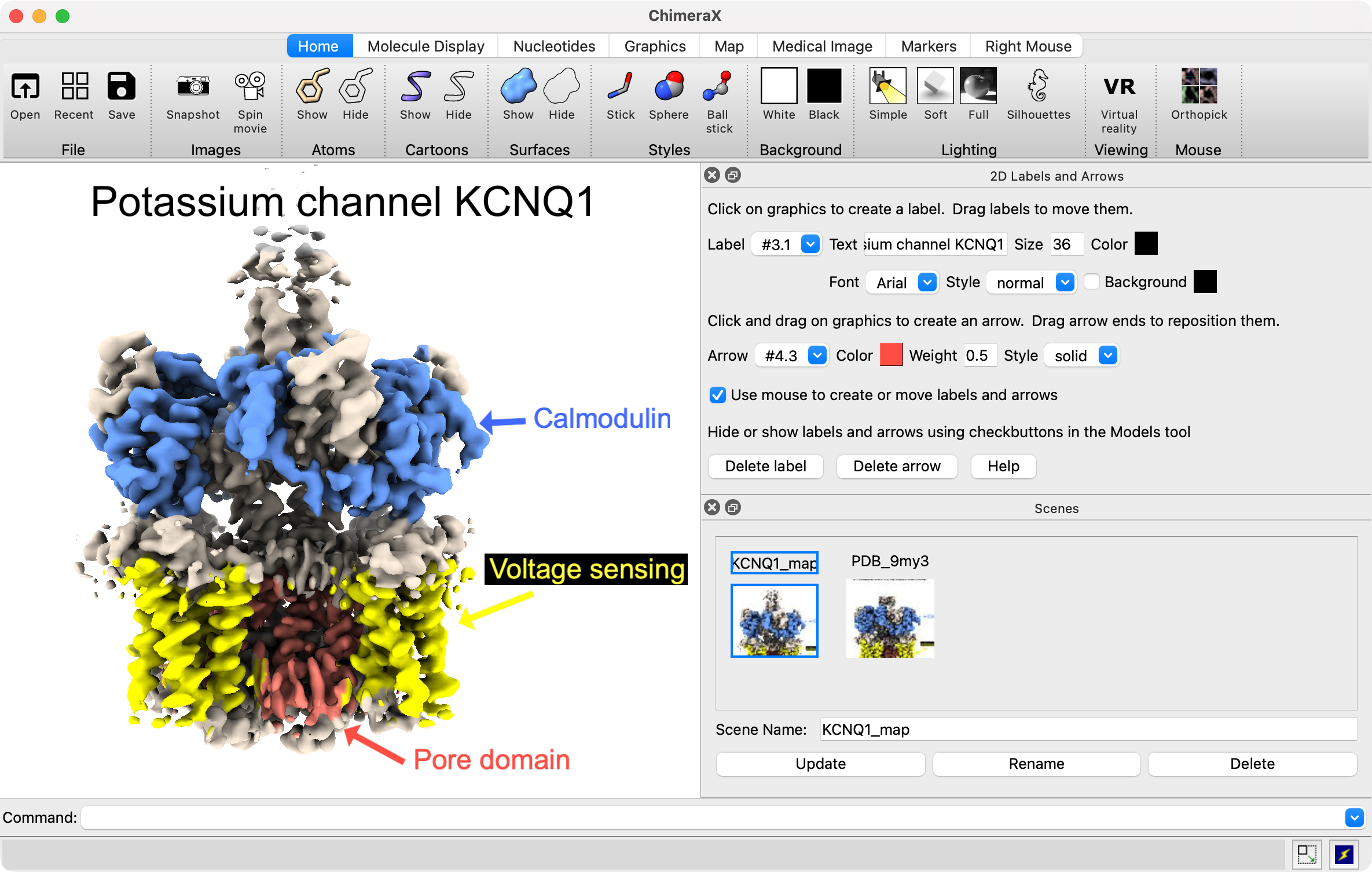
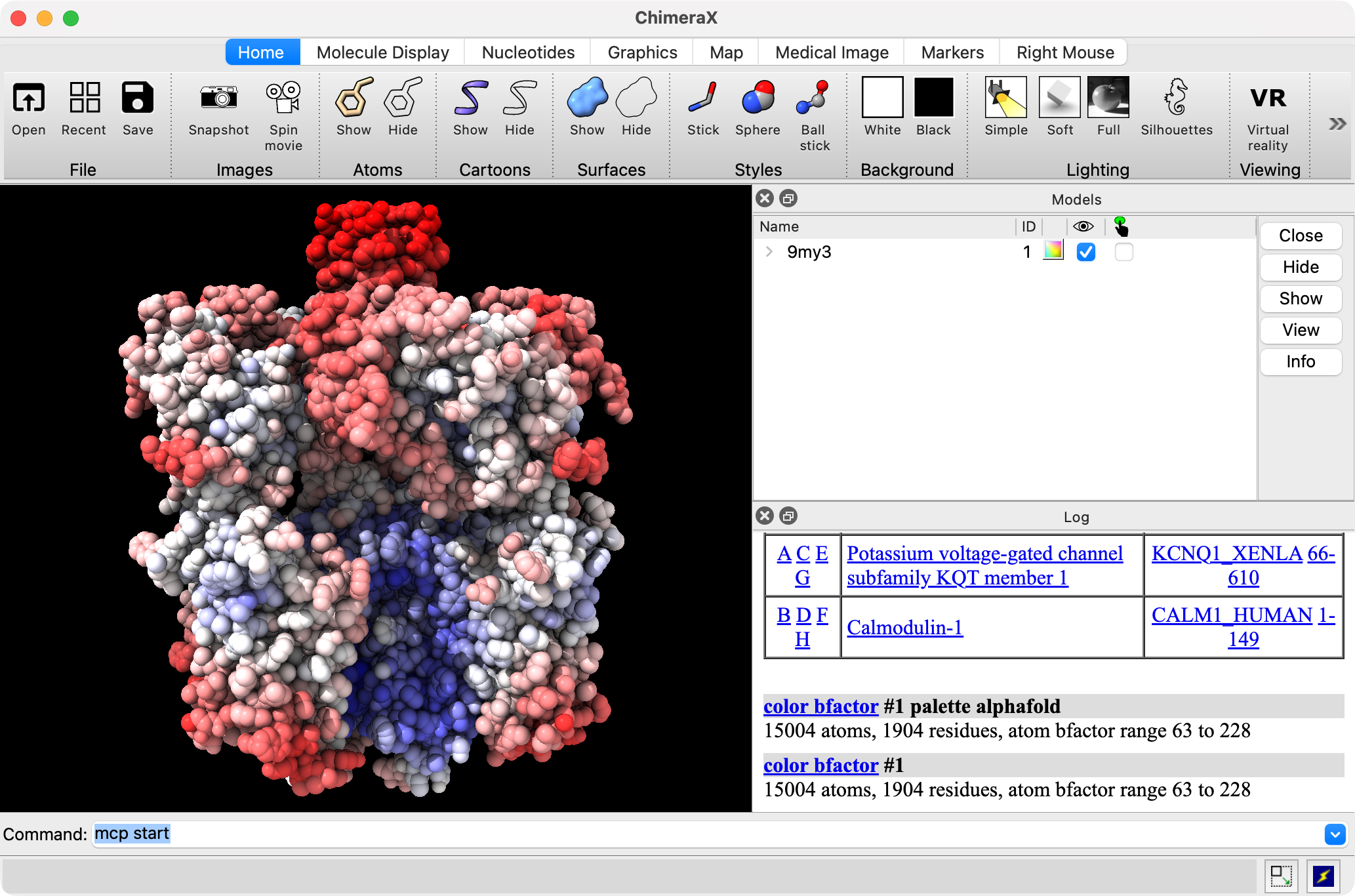
ChimeraX controlled by Claude coloring by bfactor. | 
Claude Desktop requesting it fix the bfactor coloring to not use the AlphaFold palette. |
About half the time Claude does not realize it made a mistake, and half the time it realizes it can't do something and asks the user to help.