Showing a Segmentation of Mouse Brain Electron Microscopy
Tom Goddard
June 23, 2020
The largest brain region imaged by electron microscopy to date described here
was segmented by the authors using machine learning and 4000 hours of human curation.
Dense connectomic reconstruction in layer 4 of the somatosensory cortex.
Motta A, Berning M, Boergens KM, Staffler B, Beining M, Loomba S, Hennig P, Wissler H, Helmstaedter M.
Science. 2019 Nov 29;366(6469):eaay3134. doi: 10.1126/science.aay3134. Epub 2019 Oct 24.
Here we try to look at the segmentation using ChimeraX.
Admirably, the authors made this large data set
available.
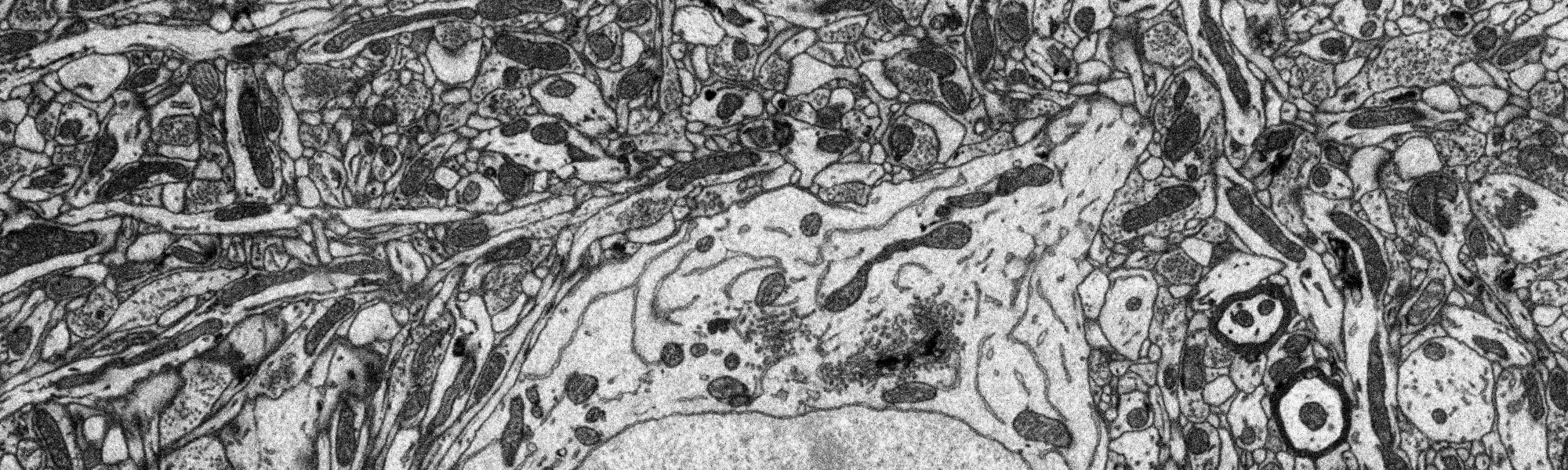
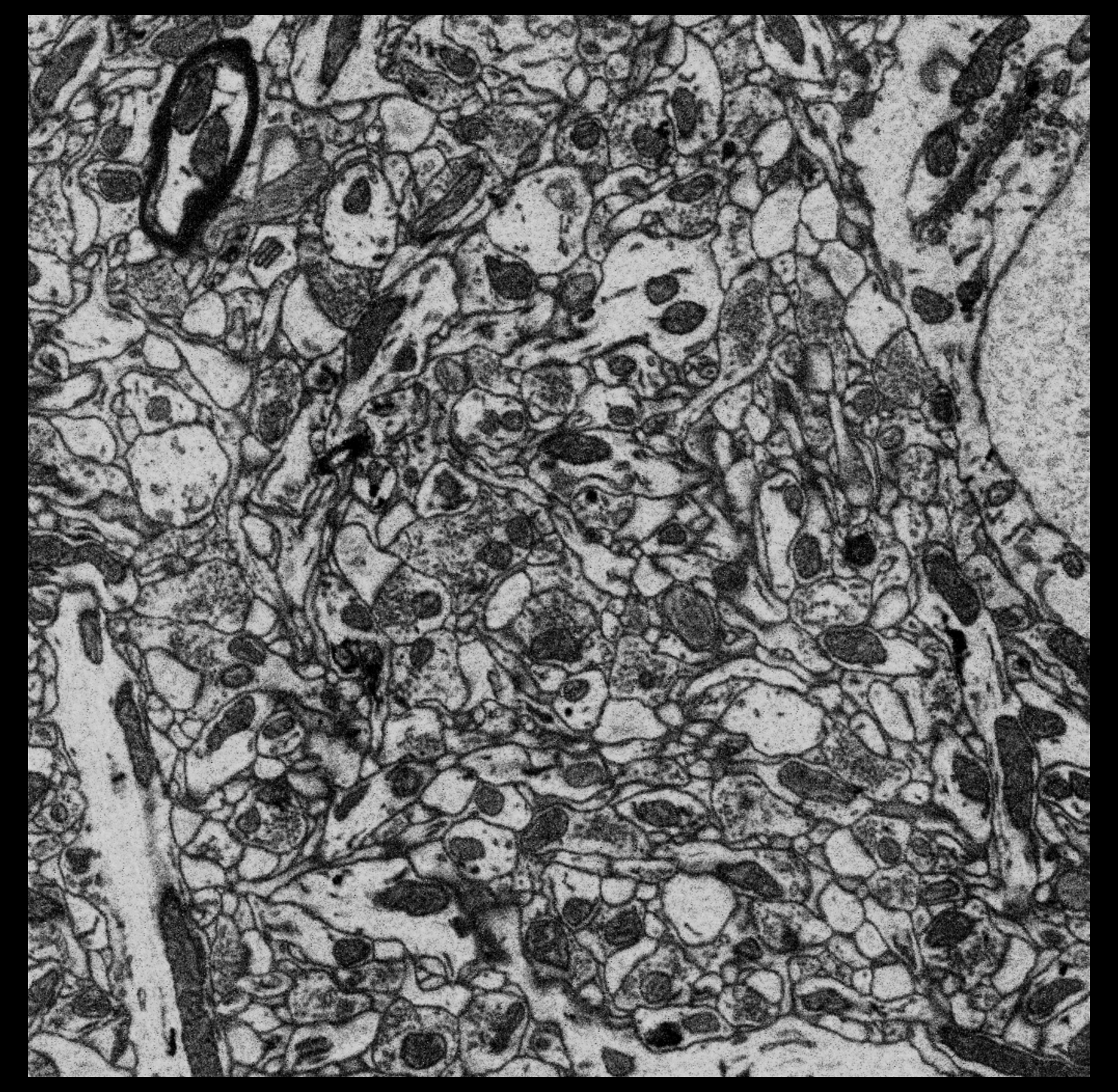
Figure 1: Small part (1/4 width, 1/10 height) of single plane of EM data.
Full plane.

| 
| 
|
Surfaces of 96 neurons with soma at least partially in volume
(video rotating and
showing each neuron).
|
Surface of one neuron (yellow) with spineheads (red)
(video slicing).
|
Small region (1/200) of volume (11x11x28 um) with
axons green, dendrites red, and spineheads blue
(video slicing).
|
open seg_step884.cmap
seg surf #1 each neuron
light soft
|
open seg_step884.cmap
seg surf #1 where neuron=1 color yellow
seg color #1 red surf #2 by spinehead
|
open image_x1y1z1.cmap
open seg_x1y1z1.cmap
hide #2
seg color #2 red map #1 by dendrite
seg color #2 lime map #1 by axon
seg color #2 blue map #1 by spinehead
volume #1 level 95,0 level 120,0.9 level 234,1
|
Figure 2:
Displaying segmentation surfaces and coloring EM image data using the
segmentation
command.
Example ChimeraX commands use the data files provided below.
Example image and segmentation files
Here are image and segmentation files assembled from the a above publications
for visualization in ChimeraX.
I am not providing the full data at full resolution because I do not have the
resources to
host hundreds of Gbytes for download. Contact me if you have a strong
need for the full resolution files, or download the data from the author's
web site and recreate the files with the Python scripts I provide below.
Movie making command scripts
Here are ChimeraX command scripts that make movies similar to those shown in Figure 2.
I try all these steps interactively, often using mouse modes and volume viewer panel
and toolbar buttons, and then I make the command script by copying commands from the
Log that were created by those user interfaces.
Data size
The imaging covers about a 0.1 millimeter cube (0.075 mm x 0.112 mm x 0.096 mm)
with voxel size 11.24 nm x 11.24 nm x 28 nm. This is about one hundredth the
diameter of a whole mouse brain (1 cm) so about 1 millionth of the brain volume.
It contains only parts of 96 neuron cell bodies (soma) which agrees with estimates
of 75 million neurons in the mouse brain. It is not large enough to contain
any complete neurons.
The image data has 6690 x 9945 x 3420 voxels, about 200 Gbytes (one byte per voxel).
It is split into 216 HDF5 files each 10243 in size. The segmentation
is similarly split into 216 HDF5 files that use compression, total size about 28 Gbytes.
The segmentation files have a 4-byte integer region id for each voxel so uncompressed
would be 4 times the size of the image data or 800 Gbytes.
Segmentation properties
The segmentation assigns id numbers to about 15 million regions (done via a
machine learning algorithm). Then individual axons, dendrites, neurons with soma
(central cell body) within the volume, spineheads, pre-synapses and post-synapses are
given separate id numbers and the regions that belong to each are specified in
additional HDF5 files.
Here are the counts of each object type:
- 15030572 regions
- 38962 axons
- 11400 dendrites
- 415797 spineheads
- 388554 synapses
- 96 neurons with soma in volume
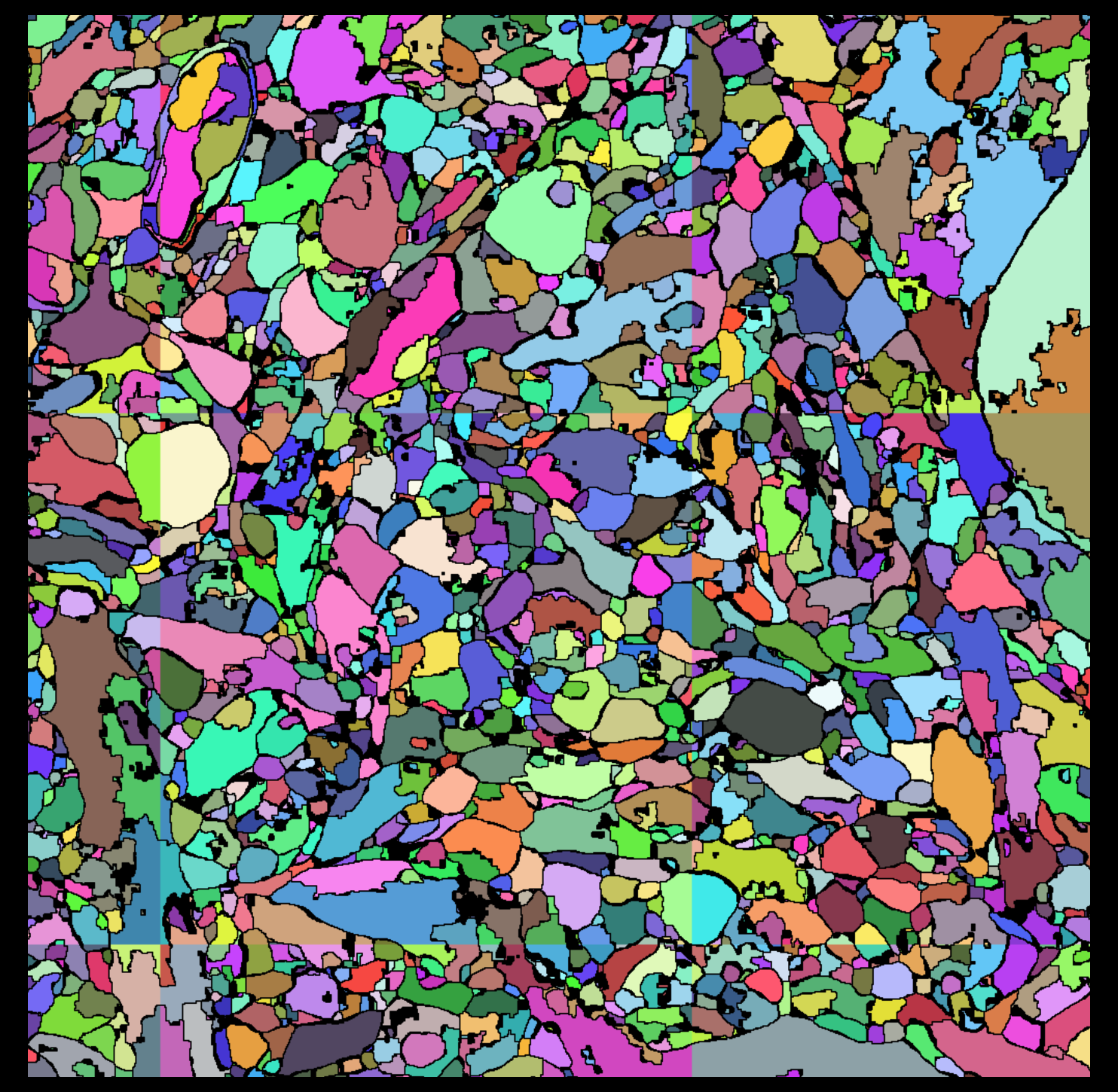
| 
|
open seg_x1y1z1.cmap
seg color #1
|
open image_x1y1z1.cmap
volume #1 level 70,0.015 level 146,0.8 level 234,1
|
Figure 3: Segmentation regions for a slice of one 10243 chunk.
There are many incorrect boundaries in the segmentation.
Simplifying data file organization
In order to be able to visualize the data I first reorganized the custom partitioning
into files used by the authors into a single file for the image data and single file
for the segmentation data so that no specialized code is needed to view it.
Single image data file.
The image data was in 216 chunks in separate files so I put it in one file.
Using 216 1 Gbyte files was no doubt easier for analysis scripts to handle.
But for visualization ChimeraX can work easily with a single file. The single
file I created is also HDF5 format although I used suffix ".cmap" (Chimera map)
and this single file contains versions of the data subsampled at many coarser
resolutions so that it can quickly display the full data, or also easily read
small chunks to show at full resolution. I used a custom script
(combine.py)
to load the 216 chunk files and copy them to a single hdf5 file.
That copy did not have the subsamples, so I opened that HDF5 file in ChimeraX
and saved as a ".cmap" file. This did not require a lot of memory (done on
my normal desktop Mac with 32 Gbytes) because ChimeraX reads only part of
the data at a time when copying. Still a 200 Gbyte file is inconvenient, so
I also made subsampled versions at step 4,4,2 (along x,y,z) and 8,8,4 for
sharing with others.
Single segmentation file.
I combined the chunked 216 segmentation files in the same way. Those files used
a slow compression method ("deflate" used by zip, 200 Mbytes/sec) which made the
conversion a slow process (several hours). The file I created uses a more
modern compression method
called blosc:lz4hc that can decompress at speeds (5-10 Gbytes/sec) faster than SSD drives
can read and is standard in HDF5 so add no additional requirements to use.
The single segmentation files is 28 Gbytes and includes subsampled copies of the
data for fast access. Compression factor is about 30.
Object ids in segmentation file.
The segmented objects (axons, dendrites, spineheads, presyanpses, postsynapses, neurons)
have their own id numbers. The authors list which region ids correspond to which
axon id, dendrite id, ... in several additional files. I included all these in
the single segmentation file each as an
integer array of length equal to the total number of regions (15030572).
The axon array lists the axon id for each of the 15 million regions.
Regions that are not axons have axon id 0.
These attributes remap region id numbers to axon id numbers, dendrite
id numbers, spinehead id numbers, ... and can be directly used with the ChimeraX
segmentation command.
HDF5 files contain an arbitrary directory structure with any number of arrays
so these attribute arrays are included under /attributes in the segmentation file.
The attributes are created with the a custom script
(object_ids.py) from the author data files
and then added to the segmentation file with the hdf5 h5copy utility as shown below.
Scripts used to simplify files
Here are the Python 3 scripts I wrote to convert the author's data files into
a single image data and single segmentation file for visualization.
To convert the authors segmentation hdf5 files to cmap format I used the following ChimeraX save command options to do the compression.
open x1y1z1.hdf5
save seg_x1y1z1.cmap region all compressMethod blosc:lz4hc compressShuffle false





