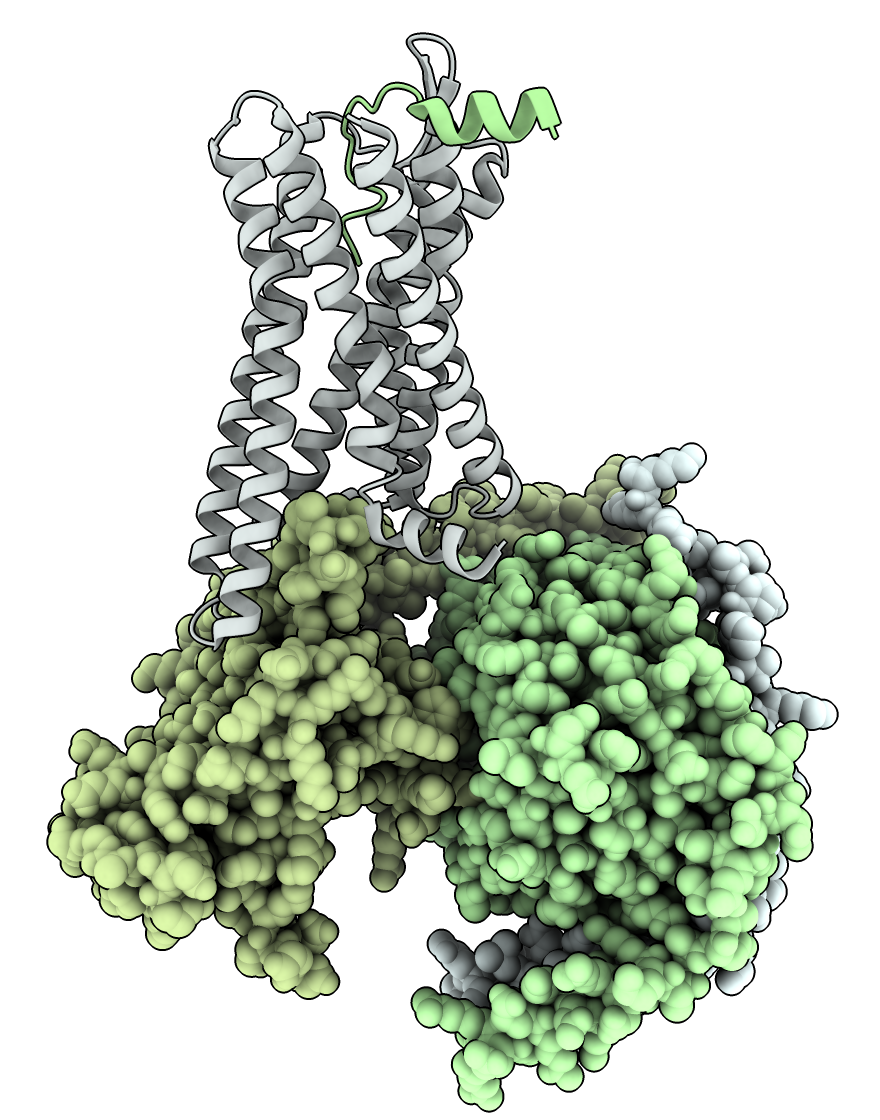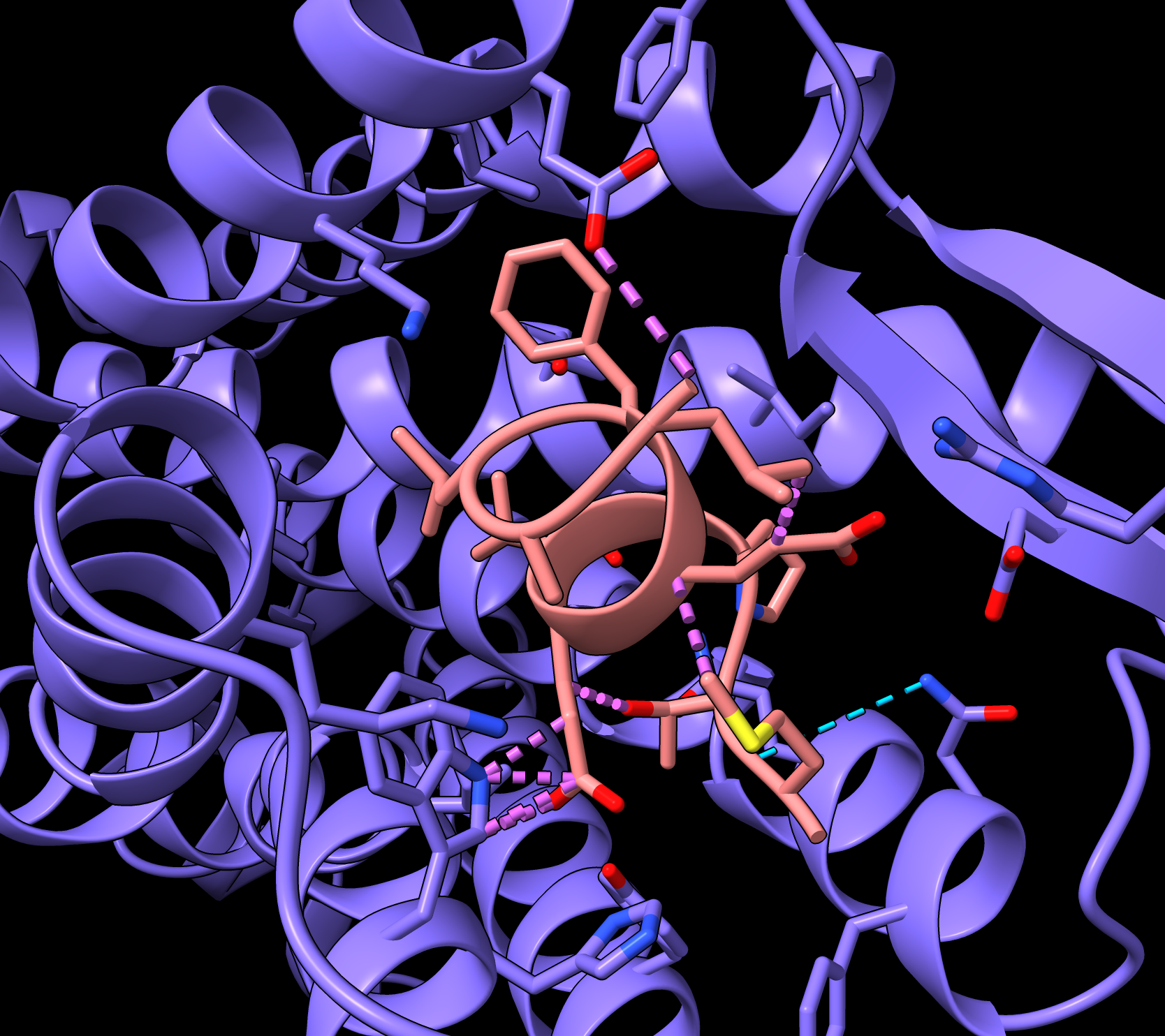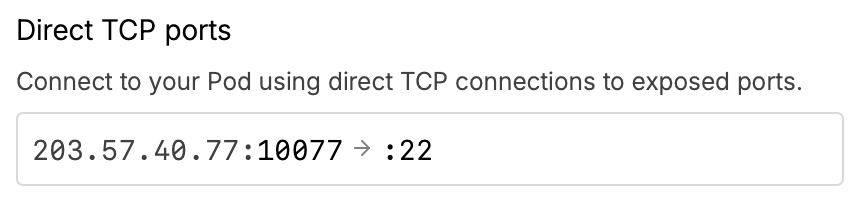
RunPod Details web page lists IP and port. |
chmod go-r ~/Downloads/id_ed25519.txt
IP=203.57.40.77 PORT=10077
scp -r -P $PORT -i ~/Downloads/id_ed25519.txt ~/Downloads/opioid_peptide root@$IP:
ssh root@$IP -p $PORT -i ~/Downloads/id_ed25519.txt
cd opioid_peptide time /workspace/boltzgen/bin/boltzgen run 8f7q.yaml --output results3 --protocol peptide-anything --num_designs 3 --budget 1 >& output3 &
tail -f output3When the timing info is output the boltzgen run has completed and you type Ctrl-C to exit the "tail -f" command which was monitoring the output file.
exit # This logs out of the VM and gets your terminal back to your laptop. scp -r -P $PORT -i ~/Downloads/id_ed25519.txt root@$IP:opioid_peptide .