Toolbar Icons
Initial Display
Manipulation
Appearance Presets
Thresholds and Windowing
Volume Rendering Options
Cropping and Clipping
Virtual Reality
Sample Data
Opening Data
Toolbar Icons
Initial Display
Manipulation
Appearance Presets
Thresholds and Windowing
Volume Rendering Options
Cropping and Clipping
Virtual Reality
Sample Data
Development of these features is supported in part by the NIAID. ChimeraX can also read the NIfTI (.nii) and NRRD (.nrrd) formats often used for medical images. See also: Download DICOM, DICOM Browser, DICOM Metadata, Segmentations, Volume Viewer, Orthoplanes, ui view, DICOM in VR tutorial, DICOM segmentation tutorial, ChimeraX for Medical Image Analysis
In ChimeraX, all of the DICOM files in a directory and its subdirectories can be opened in a single operation if the format is specified explicitly:
Menu: File... Open DICOM Folder – in the resulting dialog, browse to location, choose directory, click Choose
– or –
clickin the Medical Image tab of the Toolbar – in the resulting dialog, browse to location, choose directory, click Choose
– or –
Command: open directory-pathname format dicom
...where directory-pathname may contain * as wildcard to lessen typing, and should be enclosed in quotation marks if it includes spaces, for example:
open "~/Desktop/2-CT 5*" format dicom
Each series that is found will be opened as a separate model.
Many icons in the Toolbar across the top of the ChimeraX window are useful for working with DICOM data. The Medical Image tab contains an application-specific collection of icons:
Mouse modes are described further in the section on manipulation.
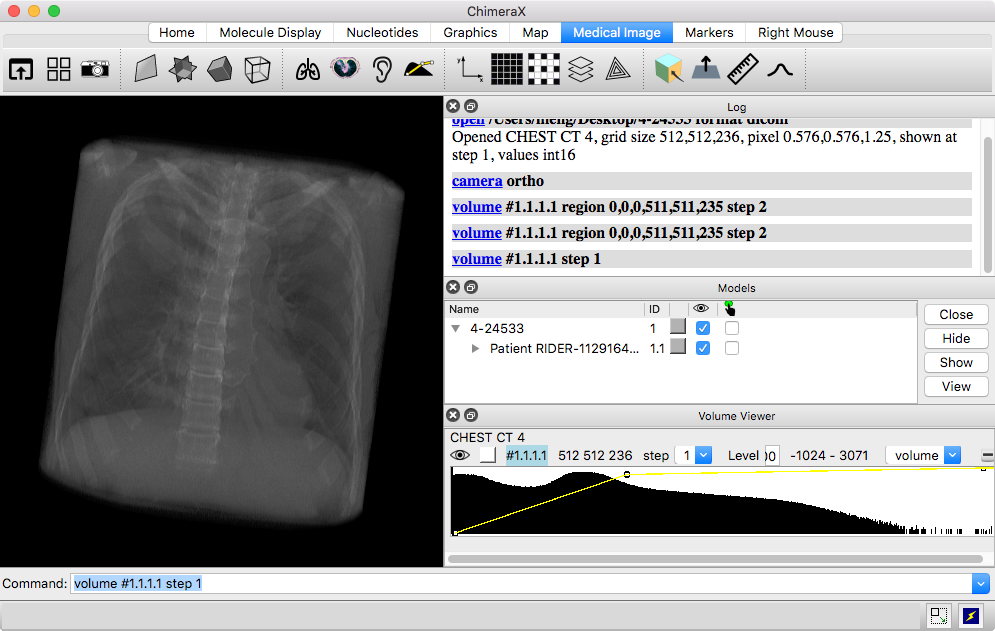
Only a single Z-plane is shown at first. The Volume Viewer tool with a histogram of the data is automatically shown, with plane as the chosen display style.
| [this dataset provided below] |
|---|
 |
For 3D volume rendering, choose volume display in Volume Viewer. This shows the full region in the transparent “image” style. The thresholds may need to be adjusted to improve the appearance after switching display styles.
For full resolution, set the step value to 1
or click the toolbar icon
![]() .
Subsampling (step > 1) decreases computational demands and
may be needed for responsive rendering of large datasets, particularly in
virtual reality. Step size may be adjusted automatically
upon initial display or when the amount of data shown is changed.
.
Subsampling (step > 1) decreases computational demands and
may be needed for responsive rendering of large datasets, particularly in
virtual reality. Step size may be adjusted automatically
upon initial display or when the amount of data shown is changed.
When multiple datasets or other models are open at the same time,
they can be hidden or shown individually, such as with the
![]() icons above their histograms in
Volume Viewer, the
icons above their histograms in
Volume Viewer, the
![]() checkboxes in the Model Panel,
or the volume command.
Many settings can be adjusted with either the
Volume Viewer graphical interface
or the volume command,
for example: volume all style image region all step 1
checkboxes in the Model Panel,
or the volume command.
Many settings can be adjusted with either the
Volume Viewer graphical interface
or the volume command,
for example: volume all style image region all step 1
Commands that are frequently useful:
For orthographic projection:
camera ortho
...or to go back to perspective projection:
camera mono
To center and scale the display to fit in the graphics window:
view
...or click graphics icon
![]()
To center and scale as above, plus reset to initial orientation:
view orient
...or click graphics icon
![]()
| [this dataset provided below] |
|---|
 |
Some DICOM files contain time series that can be played with the time slider shown automatically by ChimeraX or with the vseries command.
Some DICOM files contain SEG objects (segmentations as binary maps), and these are automatically associated with the corresponding image series to synchronize plane display. They can also be displayed as isosurfaces, for example: volume #1.1.1.1.5 show style surf level 1 region all
Segmentations described as radiotherapy structure set (RTSTRUCT) contours are also read and displayed.
Default assignments:
| rotate
|
left mouse button |
| trackpad click-drag
Mac: also trackpad 2-finger rotation if multitouch* is on |
|
| rotate VR hand controller with trigger pressed | |
| translate
|
middle mouse button |
|
Mac: trackpad + Alt,
3-finger drag if multitouch* is on
Windows: trackpad + Ctrl-Alt |
|
| move VR hand controller with trigger pressed | |
| zoom
|
mouse scroll wheel |
|
Mac: trackpad pinch or 4-finger drag if multitouch* enabled,
otherwise 2-finger drag
Windows: trackpad 2-finger drag |
|
| move VR hand controllers farther apart or closer together with both trigger buttons pressed |
Initially, the right mouse button (= trackpad + Alt on Windows,
trackpad + ![]() on Mac)
is also assigned to translate
on Mac)
is also assigned to translate
![]() ,
but can be reassigned by clicking any icon in the following tabs of the
Toolbar:
,
but can be reassigned by clicking any icon in the following tabs of the
Toolbar:
...or the last few icons in the in the Medical Image tab, as detailed above. Buttons can also be reassigned with the mousemode command.
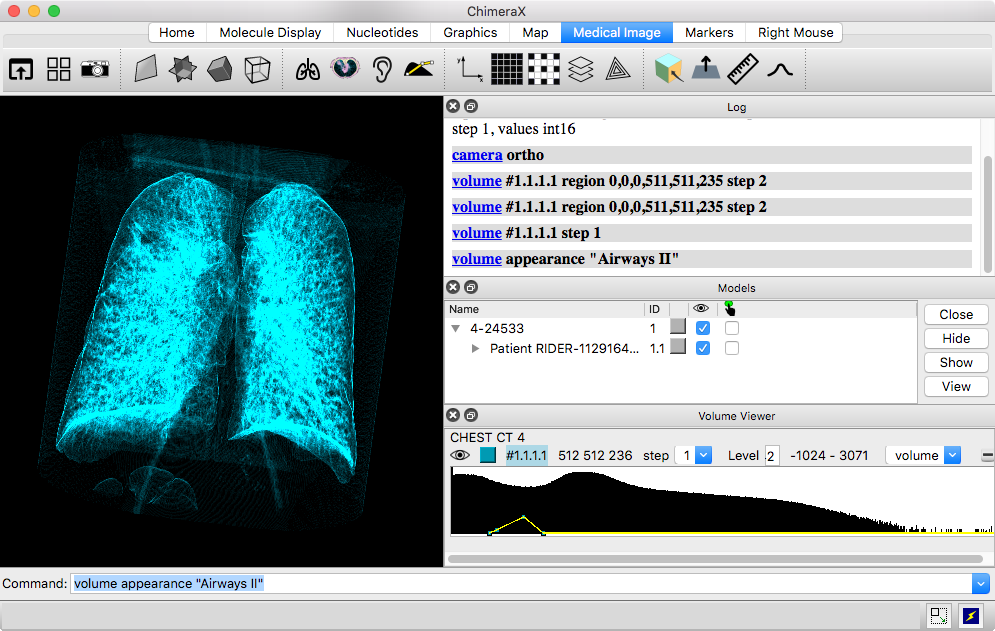
ChimeraX includes rough implementations of the Horos 3D presets and some of the MRIcroGL color lookup tables. They can be applied by name with the volume command appearance option, for example:
volume #1 appearance "Airways II"
volume #1 appearance CT_Muscles
The implementations are only approximate, as there may not be a direct correspondence between settings in different programs, and parts of the definition files may have been ignored.
The following presets, also available as toolbar icons, are defined in ChimeraX only:
Users can also define their own custom appearance presets.
| [this dataset provided below] |
|---|
 |
Thresholds are the control points on the histogram for mapping values to colors and intensities. They can be moved by dragging in the Volume Viewer histogram, and added and deleted using the context menu (right-click or Ctrl-click depending on platform). The color of the most recently clicked threshold can be changed by clicking the color square above the left end of the histogram.
Applying a 3D preset sets thresholds and colors, among other things. Threshold levels and colors can also be set explicitly with the volume command, for example:
volume #1 level -742,0.0 level -683,0.049 level -481,0.25 level -333,0.0 color 0,61,71 color 0,61,71 color 0,61,71 color 0,61,71
A windowing mouse mode allows adjusting image display thresholds collectively:
| windowing
|
vertical motion adjusts window level (moves all thresholds in parallel to higher or lower values), horizontal motion adjusts window width (moves thresholds symmetrically farther apart or closer together), and the dominant type of motion wins out; for adjusting multiple windows within a single histogram, see details |
A mouse mode can be assigned to a button as described above.
Additional volume command options that may be useful:
Cropping limits the display of a dataset to a smaller rectangular box or slab, making it easier to see features of interest while reducing the computational demands of rendering. Cropping can be done with:
A cropped slab display can be moved along an axis using
the move planes mouse mode
![]() .
Single planes can be viewed as described above.
.
Single planes can be viewed as described above.
In contrast, clipping may not have much effect on rendering speed, since calculations are still performed on the hidden parts. ChimeraX has near/far clipping planes that always face the viewer, plus the ability to clip/slab at any angle (so-called front/back planes). Both kinds can be controlled with the clip command or with the following mouse modes:
| clip
|
activate and translate front clipping plane, +Alt back plane, +Shift slab (front and back in same direction), +Alt-Shift slab thickness (front and back in opposite directions); if only near/far (fixed-orientation) planes already active, move those instead of front/back (rotatable) planes |
| clip rotate
|
activate and rotate front/back clipping planes |
A mouse mode can be assigned to a button as described above.
The near/far planes can also be manipulated in the
Side View
(menu: Tools... General... Side View).
To turn off all clipping: clip off
or ~clip
DICOM data can be viewed in ChimeraX virtual reality. Important aspects of using VR:
Clicking a mouse mode icon
with a hand controller assigns
whichever button was clicked to the corresponding function.
In VR, leaving the triggers with their
default assignments (rotation, translation,
zooming) is recommended, but up to four other functions can be assigned,
to the grip and trackpad buttons on each controller.
Useful modes for DICOM in VR include
move planes
![]() ,
crop volume
,
crop volume
![]() ,
and windowing
,
and windowing
![]() .
.
See also: DICOM in VR tutorial, DICOM segmentation tutorial, the VR at UCSF website, VR setup advice and observations on different systems: Windows, Mac, Linux
4-24533.zip – a DICOM stack representing one CT scan of many in the RIDER Lung CT collection from The Cancer Imaging Archive (TCIA):
The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Clark K, Vendt B, Smith K, et al. Journal of Digital Imaging. 2013;26(6):1045-1057.Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Zhao B, James LP, Moskowitz CS, Guo P, Ginsberg MS, Lefkowitz RA, Qin Y, Riely GJ, Kris MG, Schwartz LH. Radiology. 2009 Jul;252(1):263-72.
Zhao B, Schwartz LH, Kris, MG. (2015). Data From RIDER Lung CT. The Cancer Imaging Archive. doi: 10.7937/K9/TCIA.2015.U1X8A5NR
The data have been anonymized and are freely available. The folder should be unzipped and the files opened in ChimeraX as described above. A significant feature of this scan is a tumor in the right lung.
Note that the Download DICOM tool makes it easy to browse and download TCIA datasets from within ChimeraX.