Opened 3 years ago
Closed 3 years ago
#8388 closed defect (fixed)
gaps between pseudobonds and ribbons in printable representations
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Depiction | Version: | |
| Keywords: | Cc: | Tom Goddard, phil.cruz@…, Elaine Meng | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description (last modified by )
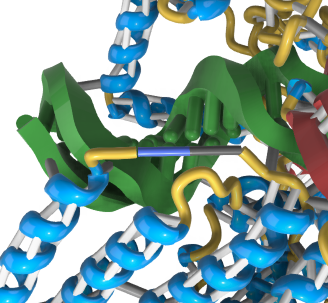
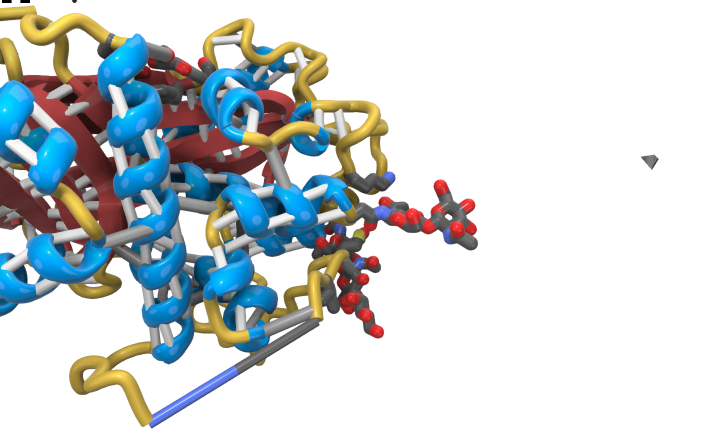
We're noticing that the pseudobonds are not well connected to the ribbons, so aren't going to be super stable when printed. For example: [cid:image001.png@01D9349A.CF93ACF0][cid:image002.png@01D9349A.CF93ACF0] This is from PDB 1A36. Similar in: [cid:image003.png@01D9349A.CF93ACF0] From PDB 1T1F. (This one also had some random debris off to the right. Not sure what is up there? It's in all the print representations. Phil didn't see it using ChimeraX with the presets alone. We will rerun through the workflows with the newer daily build when Michal updates it tomorrow just in case.) Wondering what your thoughts might be on the bond issue. I was thinking maybe need to just make them a bit longer or ball-cap them to create better overlaps, but depending on the angle, this might not fix the issue. Thanks, K Kristen Browne, MSc, MscBMC Contractor - MSC, Inc. | A Guidehouse Company 3D Modeling and Biovisualization Specialist Bioinformatics and Computational Biosciences Branch (BCBB)<https://www.niaid.nih.gov/research/bioinformatics-computational-biosciences-branch> OCICB/OSMO/OD/NIAID/NIH 5601 Fishers Lane, Room 4A60 Rockville, MD 20852 Office 202-253-5228
Attachments (5)
Change History (18)
by , 3 years ago
| Attachment: | image001.png added |
|---|
comment:1 by , 3 years ago
| Cc: | added |
|---|---|
| Component: | Unassigned → Depiction |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
comment:2 by , 3 years ago
I can certainly see a gap in 1a36 between the missing-structure pseudobond and the ribbon using the "monochrome printable" preset.
comment:3 by , 3 years ago
| Cc: | added; removed |
|---|---|
| Owner: | changed from to |
When the atoms for pseudobond end points, in this case C and N backbone atoms, are not shown and the ribbon is shown, the pseudobond end points are placed on the ribbon. For C and N atoms the positions are placed 1/3 of the way from the CA atom toward the next residue. As seen in the examples in this ticket that is sometimes a tiny bit past at the end of the ribbon. That is pretty reasonable since the C and N atoms are the terminal atoms of the ribbon for a missing segment.
There are a large number of hacks I can think of to make the missing pseudobond connect better to the ribbon without a gap. All of them involve adding code to the NIH 3D scripts. I believe the current ChimeraX behavior is reasonable, and this 3D printing case is a very special one.
Here are a range of methods to make this more suitable for 3D printing.
1) Don't show the missing segment pseudobonds. The segments of the structure will be connected by struts so they are not needed.
2) Show the C and N atoms and CA atoms of the residues for the missing segement. I've attached an image showing what this looks like. The CA may not be positioned on the ribbon (it is for alpha helix) so this may not work all the time. Maybe the CA is in general close enough to overlap the ribbon.
3) Replace the pseudobonds with new pseudobonds that go to the CA atoms.
4) Set the effective atom coordinate for the C and N atoms on the ribbon to the CA effective atom coordinate for just the missing segment end residues (using Atom.set_ribbon_coord).
5) Make no changes. If printed some missing pseudobonds may be disconnected but struts hold the rest together.
6) Fatten the missing segment pseudobonds. This doesn't make them longer so it doesn't make a connection when the ribbon is colinear with the pseudobond.
I've attached a picture of a missing structure pseudobond for 1a36 that does not connect strongly to the ribbon, and another picture of one work-around which is to show the C and N atoms that the pseudobond joins and their CA atoms.
by , 3 years ago
| Attachment: | 1a36_gap.png added |
|---|
1a36 missing segment pseudobond not connected to ribbon.
by , 3 years ago
| Attachment: | 1a36_atoms.png added |
|---|
1a36 missing segment pseudobond with C,N and CA atoms shown to make stronger connection to ribbon.
comment:4 by , 3 years ago
| Status: | assigned → feedback |
|---|
So, a couple of things. What does "1/3 of the way...toward the next residue" even mean when the next residue is missing? Shouldn't this interpolation always be constrained to the end of the ribbon?
If this depiction problem isn't going to be fixed directly, I think the NIAID folks need to pick one of those six options. I can implement whatever gets picked.
comment:5 by , 3 years ago
The ChimeraX ribbon cubic spline uses the C-alpha atoms. But it extends beyond the end C-alphas by 30% (0.3) by extrapolating. The C, N atom effective positions on the ribbon are not clamped to the end of the ribbon and are 1/3 = 0.333 of the way past the C-alpha, at the end it is done again by extrapolating the spline beyond the final C-alpha position. So this is why the N and C effective ribbon positions end up just beyond the end (0.33 position, vs 0.3 end). If I change the C++ code to extend the ribbon by 0.4 residue spacings beyond the C-alpha then the pseudobond ends inside the ribbon. I guess the 0.3 extension was chosen because the extrapolations are not going to have the right curvature since the next C-alpha is missing. So we extend minimally. I don't think this very obscure use case of 3D printing merits changing that extension parameter. The extension amount is buried in C++ ribbon code and would take some work to expose, and I do not think that is a good solution for this problem. I think showing the terminal CA-C or N-CA at missing segments or just not showing the missing segments for 3D printing are the most sensible solutions. But I agree the NIAID people should decide.
comment:6 by , 3 years ago
| Cc: | added |
|---|
So I had hoped to discuss this at today's meeting since it's a wee bit of a discussion topic as to how pseudobonds should behave in this scenario, but Kristen wasn't there. Perhaps at next week's meeting if time allows...
comment:7 by , 3 years ago
Please discuss with Phil as he actually reported the issue and can decide what is the acceptable outcome.
Get Outlook for iOS<https://aka.ms/o0ukef>
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Thursday, February 2, 2023 5:36:51 PM
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Browne, Kristen (NIH/NIAID) [C] <kristen.browne@nih.gov>; meng@cgl.ucsf.edu <meng@cgl.ucsf.edu>; pett@cgl.ucsf.edu <pett@cgl.ucsf.edu>; Cruz, Phil (NIH/NIAID) [C] <phil.cruz@nih.gov>
Subject: [EXTERNAL] Re: [ChimeraX] #8388: gaps between pseudobonds and ribbons in printable representations
#8388: gaps between pseudobonds and ribbons in printable representations
---------------------------------------+----------------------
Reporter: kristen.browne@… | Owner: pett
Type: defect | Status: feedback
Priority: normal | Milestone:
Component: Depiction | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
---------------------------------------+----------------------
Changes (by pett):
* cc: phil.cruz@…, Elaine Meng (added)
Comment:
So I had hoped to discuss this at today's meeting since it's a wee bit of
a discussion topic as to how pseudobonds should behave in this scenario,
but Kristen wasn't there. Perhaps at next week's meeting if time
allows...
--
Ticket URL: <https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F8388%23comment%3A6&data=05%7C01%7Ckristen.browne%40nih.gov%7C3d40596c89c0419b57b408db056dfc3e%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638109742180720579%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=bexwwGAXWbkHTQJVqGCnmnV5fazbrJ%2FHsL2j8Da6xQ0%3D&reserved=0>
ChimeraX <https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C01%7Ckristen.browne%40nih.gov%7C3d40596c89c0419b57b408db056dfc3e%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638109742180720579%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=LpnSfCEjpROw8xTjqjkiQqgXO9SFUV5PBF67J9oH9rQ%3D&reserved=0>
ChimeraX Issue Tracker
CAUTION: This email originated from outside of the organization. Do not click links or open attachments unless you recognize the sender and are confident the content is safe.
comment:8 by , 3 years ago
| Description: | modified (diff) |
|---|
<poke!> This is ticket about the gaps between pseudobonds and ribbons
comment:9 by , 3 years ago
| Status: | feedback → accepted |
|---|
Phil's feedback that didn't make it into the ticket during the server-upgrade chaos:
Finally had a chance to look into this. Tom gave several proposed hacks to address this, but as he said, “this 3D printing case is a very special one.”
I think the best straightforward solution is Tom’s suggestion #3 (or the similar #4, whichever is easier), i.e., make the pseudobonds go between CA atoms. I made a psdudobond file for 1a36 to test this, and the image after doing this is attached. This approach also looks ok with 1t1f.
I think this will give the best results when printing. These missing-residue pseudobonds often provide the prints mechanical stability where they most need it, and at least in these two cases the pseudocbond connections to the ribbon look quite solid, but I realize that this may not be the case with all geometries.
Thoughts?
follow-up: 5 comment:10 by , 3 years ago
I can try adding this behavior to the presets and see how we like it.
comment:11 by , 3 years ago
I have pushed an updated bundle to the toolshed that makes the missing-structure pseudobonds go between the CA atoms. I not sure what color you want these pseudobonds to be, but right now they are yellow, which is the default color for pseudobonds read from a file. Yellow seems okay to me -- it draws attention to the fact that there is missing structure (rather than looking like just another strut) without being too obnoxious. Nonetheless, the presets are for you so if you prefer another color let me know.
follow-up: 7 comment:12 by , 3 years ago
The bundle I submitted 6 days ago had a bug that prevented printable presets from working. I have just uploaded a fixed version.
comment:13 by , 3 years ago
| Resolution: | → fixed |
|---|---|
| Status: | accepted → closed |
The CA-to-CA missing-structure pseudobonds seem to have been acceptable

Added by email2trac