Work in progress January 18, 2024 - May 15, 2024
Tom Goddard
May 16 and 23, 2024
Group meeting
Topics
- Collaborative projects
- AlphaFold 3
- New ChimeraX capabilities
- Virtual reality
- Vacation photos
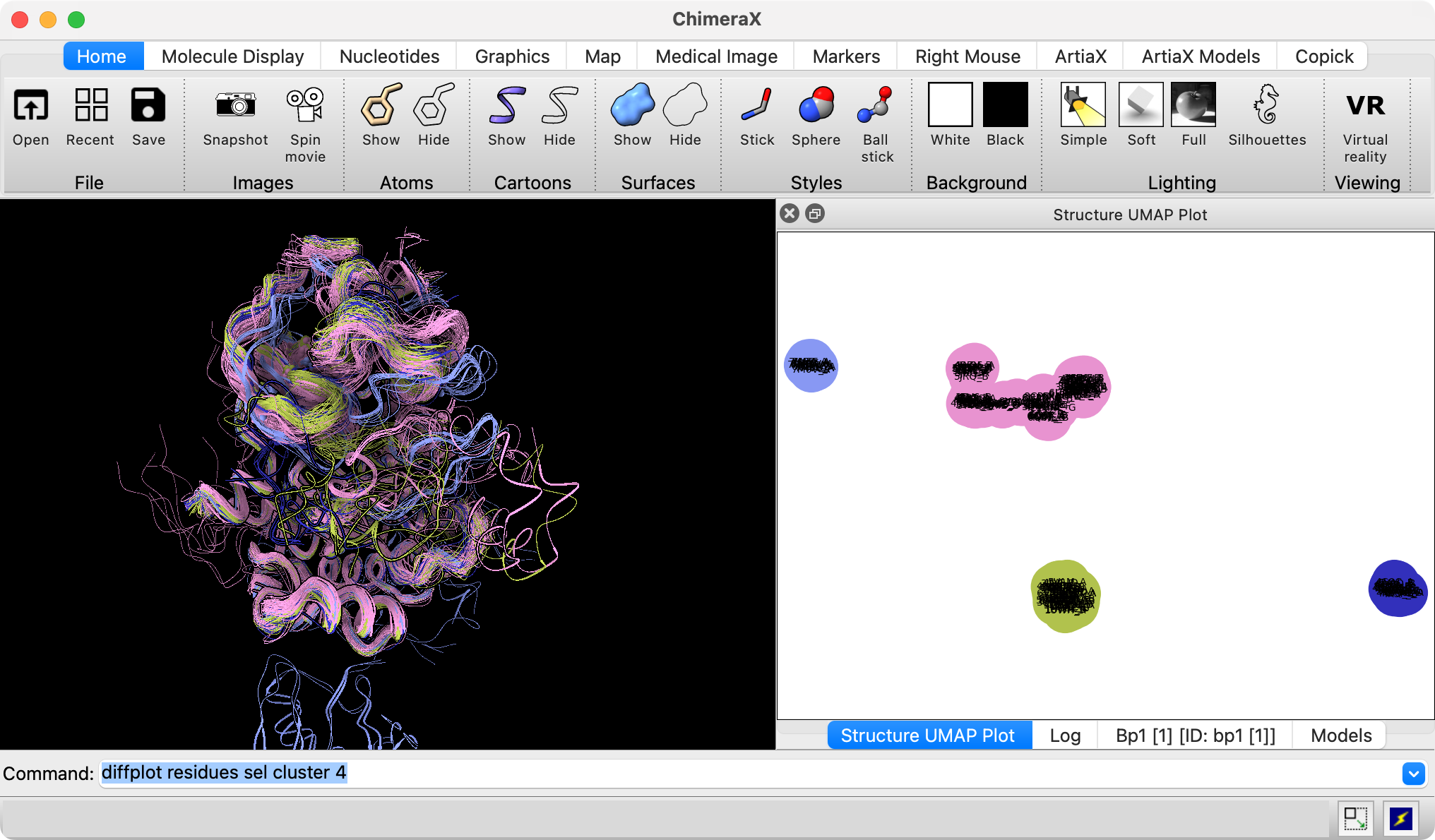
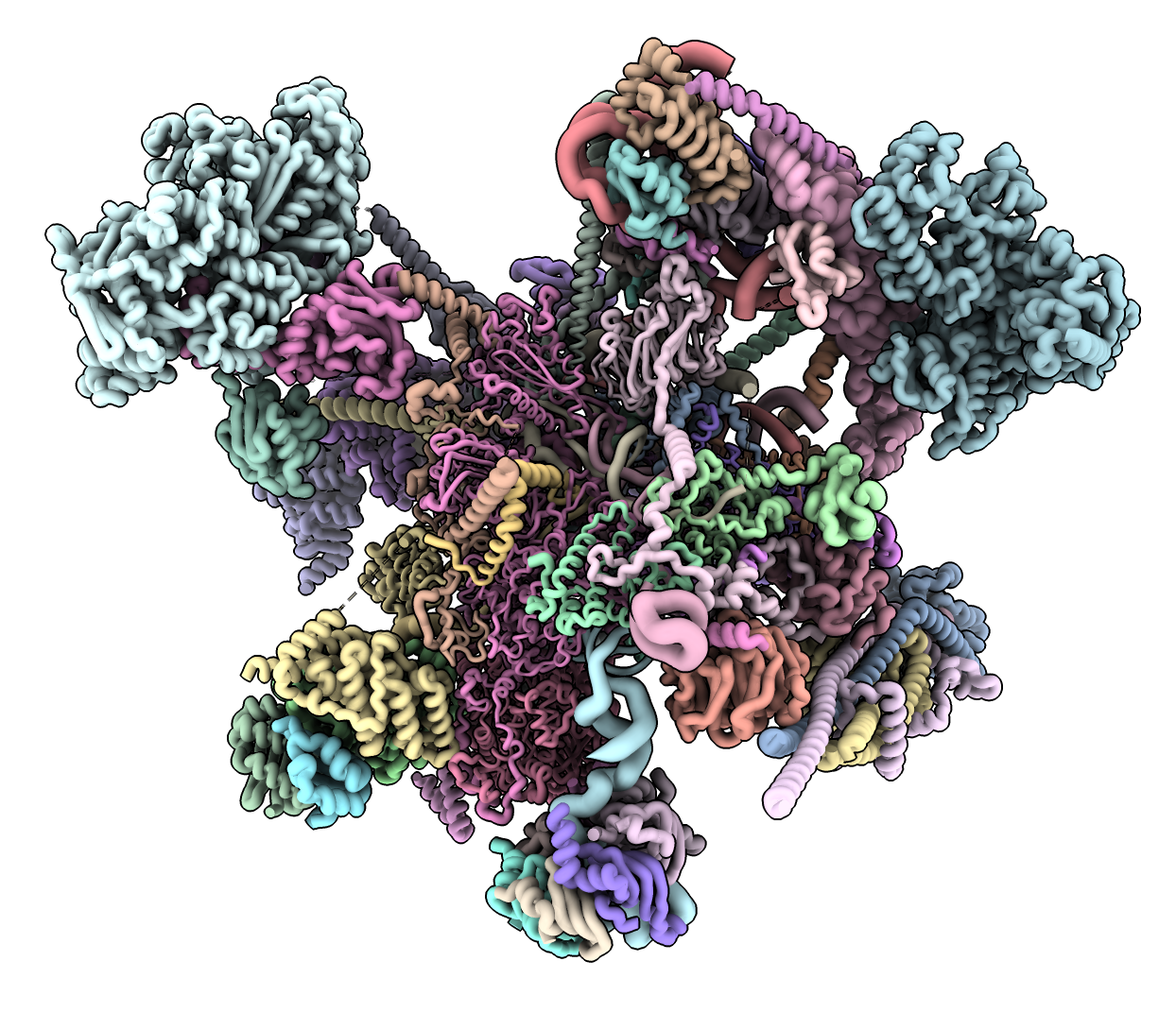
Structure conformation comparison using UMAP with Klim Verba
- Added ChimeraX diffplot command that projects selected residue coordinates to 2D using Uniform Manifold Approximation and Projection (UMAP) to help find different conformations among hundreds or thousands of structures.
- Tried on 200 B-Raf kinase structures with Klim Verba and Katrina Black (image at right).
- Distinguished different positions of key activation helix of B-Raf.
- Also tried on 200 out of 4000 AlphaFold database structures with sequences similar to human B-Raf.
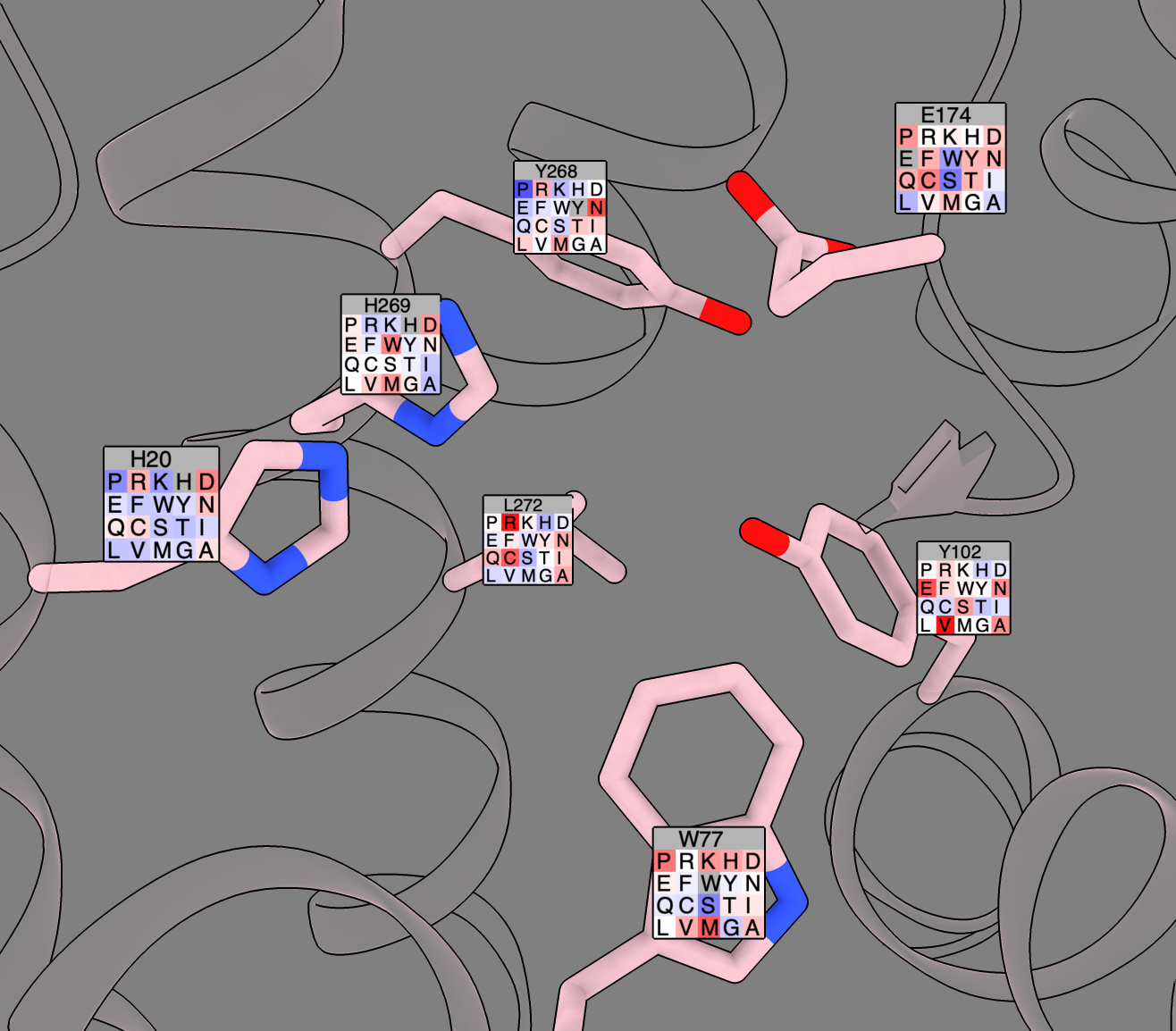
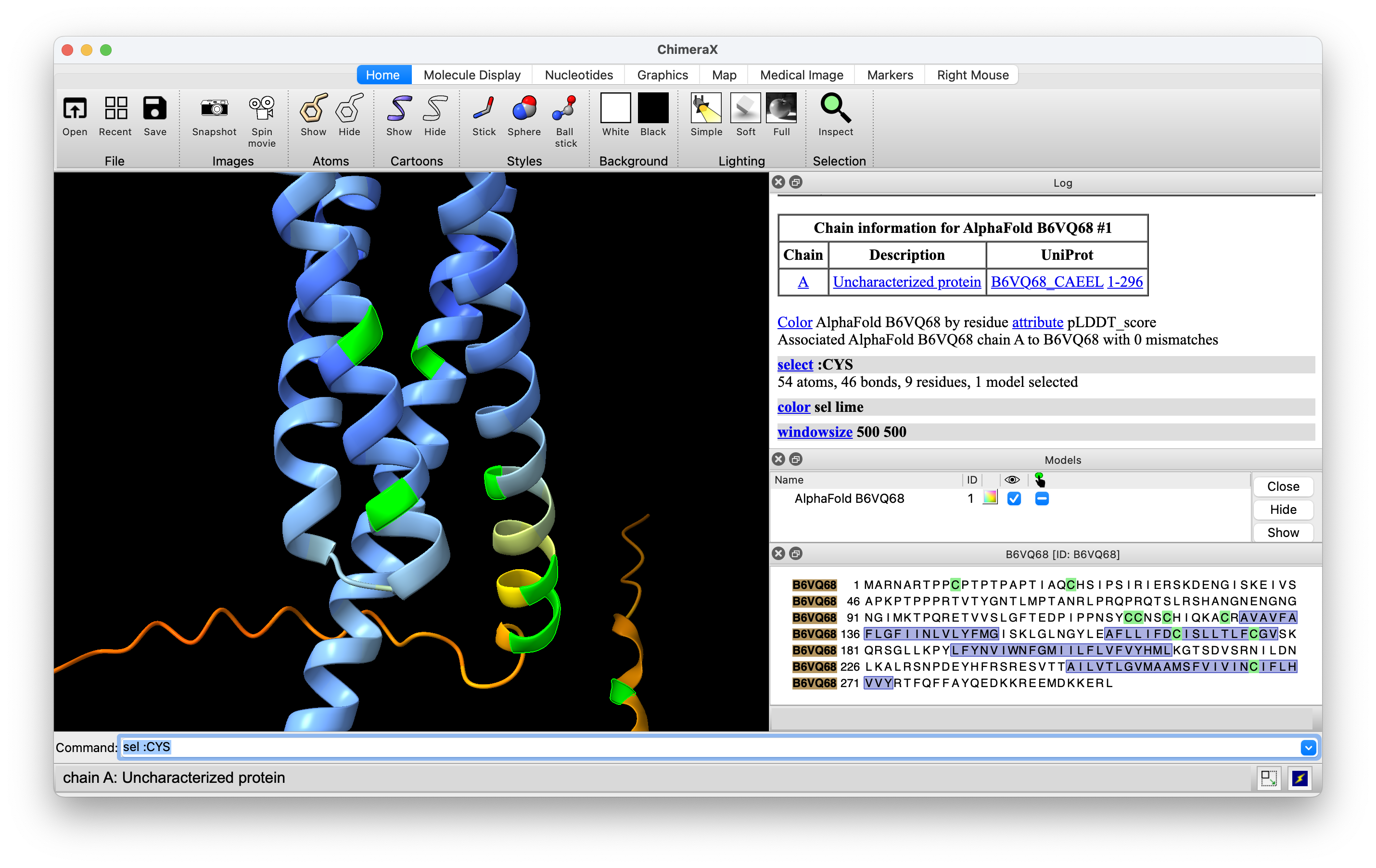
Deep Mutational Scanning visualization with Willow Coyote-Maestas and Aashish Manglik labs
- Deep mutational scanning assays a protein function for every possible single mutation of every residue.
- Aashish asked for a way to see this data on the structure.
- Tried adding fancy residue labels that show color-coded loss (red) of gain (blue) of function for each mutation
(image at right).
- Willow and Aashish are looking at GPCRs that sense extracellular pH to regulate human body pH precisely to 7.4 as described in biorxiv preprint Molecular basis of proton-sensing by G protein-coupled receptors".
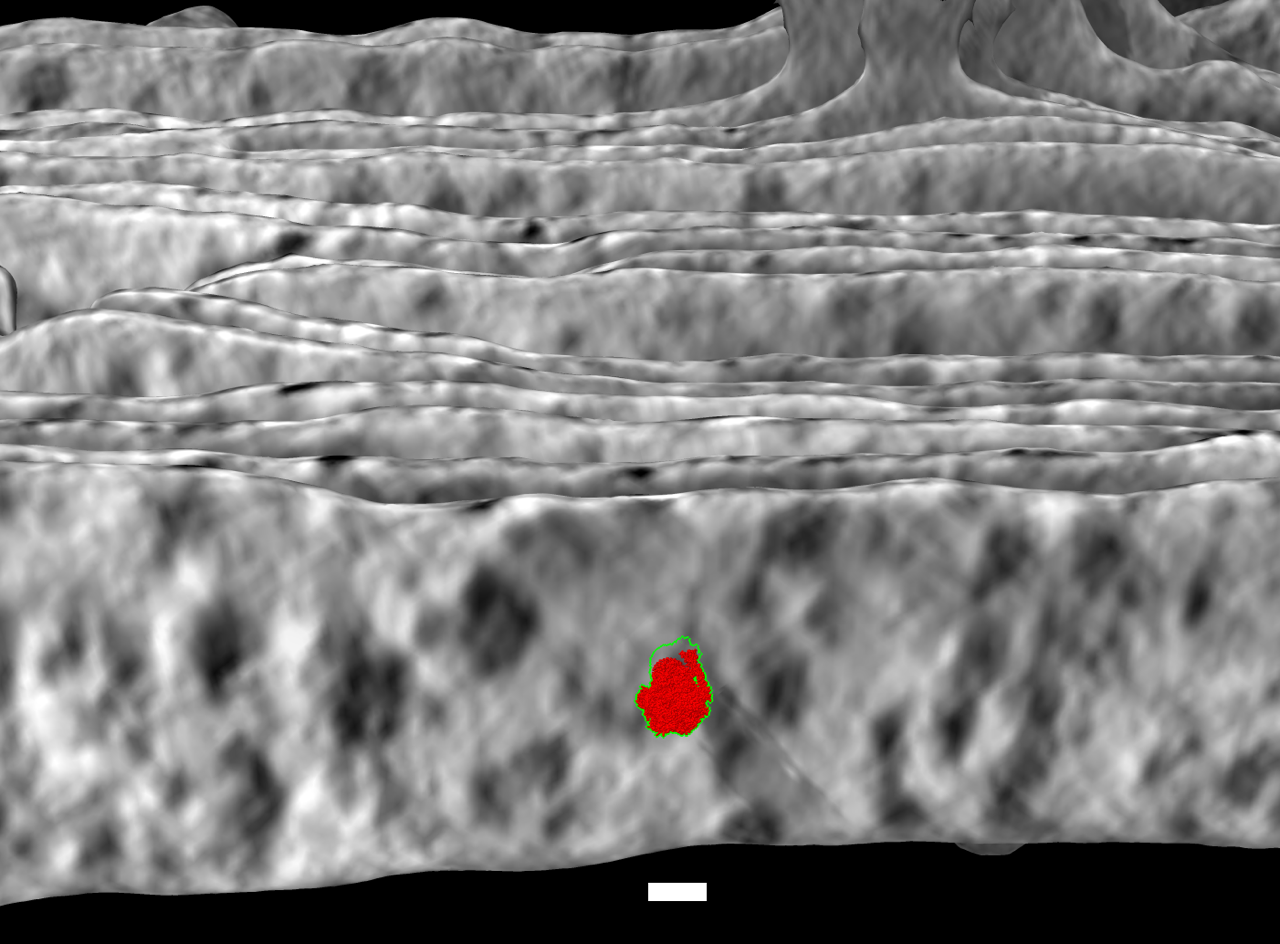
Visit to Chan Zuckerberg Imaging Institute
- Visited Utz Ermel (ArtiaX plugin developer), Shawn Zheng and Bridget Carragher at CZII in Redwood City.
- Institute goal is to reveal molecular organization inside cells by advancing electron tomography hardware and software.
- Discussed how to visualize membrane proteins such as V-ATPase on lysosomes.
- Added new capability to project a slab of density onto a segmented cellular membrane (image at right showing chloroplast thylakoid membrane with ATP synthase model in red).
- Tried machine learning membrain-seg software on minsky (used in image at right).
- Helped Utz at ChimeraX keyboard shortcuts for tomogram particle picking, cheered on by Bridget.
- Shawn Zheng visited me at UCSF a week later to discuss mouse mode to find membrane proteins that will project
density along electron beam axis to avoid missing wedge artifacts.
NIAID ChimeraX Workshop, February 2024
- Workshop hosted by Meghan, Phil, Darell, Kristen, Victor and NIAID BioViz Lab.
- Gave 2 hour ChimeraX intro class that turned into a 90 minute question and answer session plus 30 minutes of cryoEM tutorial.
- Fixed VR skyline artifacts with nvidia 4070 by disabling async spacewarp.
- Discussed ideas for wall touch screen ChimeraX improvements.
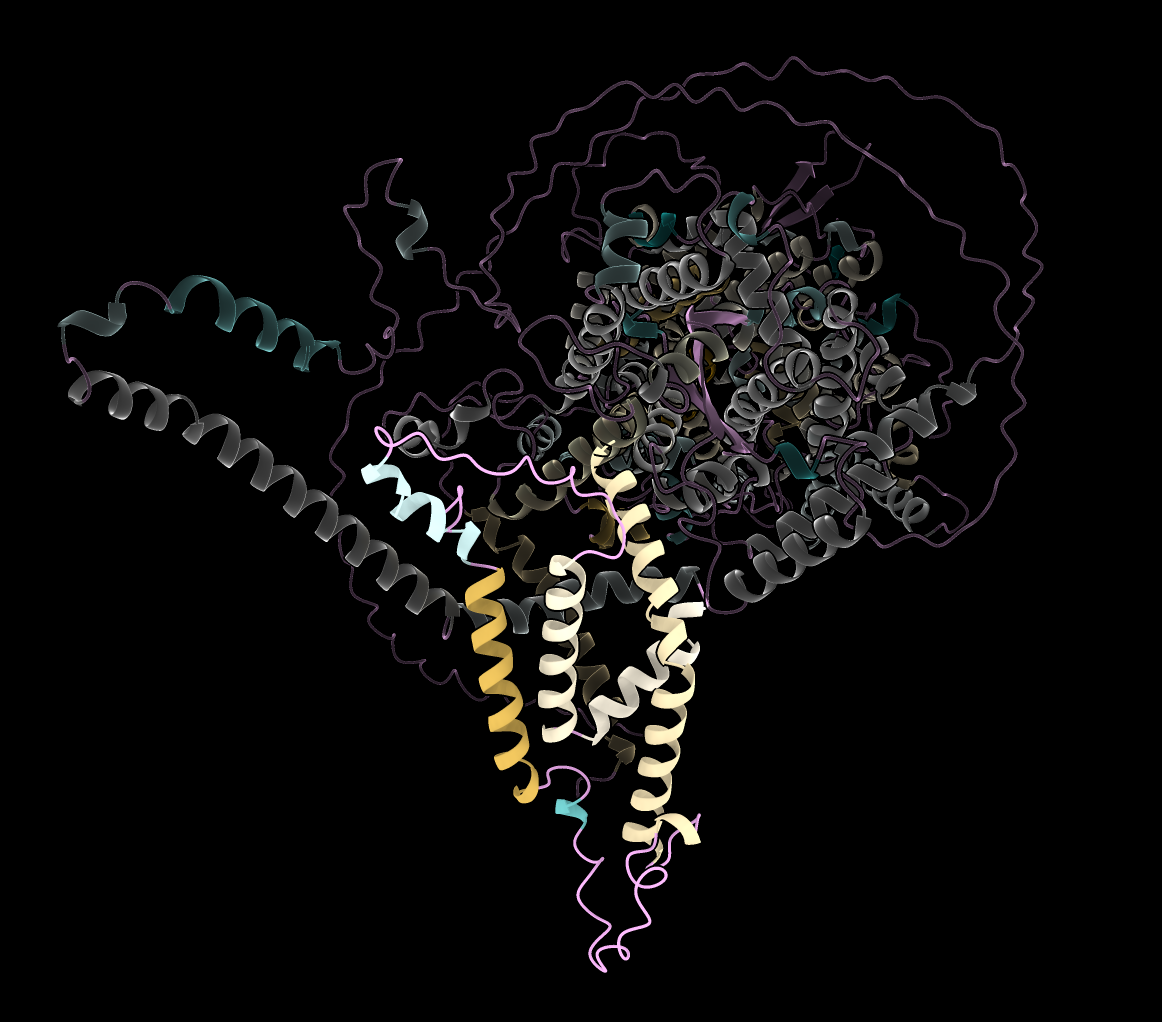
- Tried modeling malaria nutrient membrane portal complex using AlphaFold for Sanjay Desai. Only experimental structure is for a soluble form of complex before it is inserted into membrane. Wrote code to color alpha helices by hydrophobic exposure (image at right).
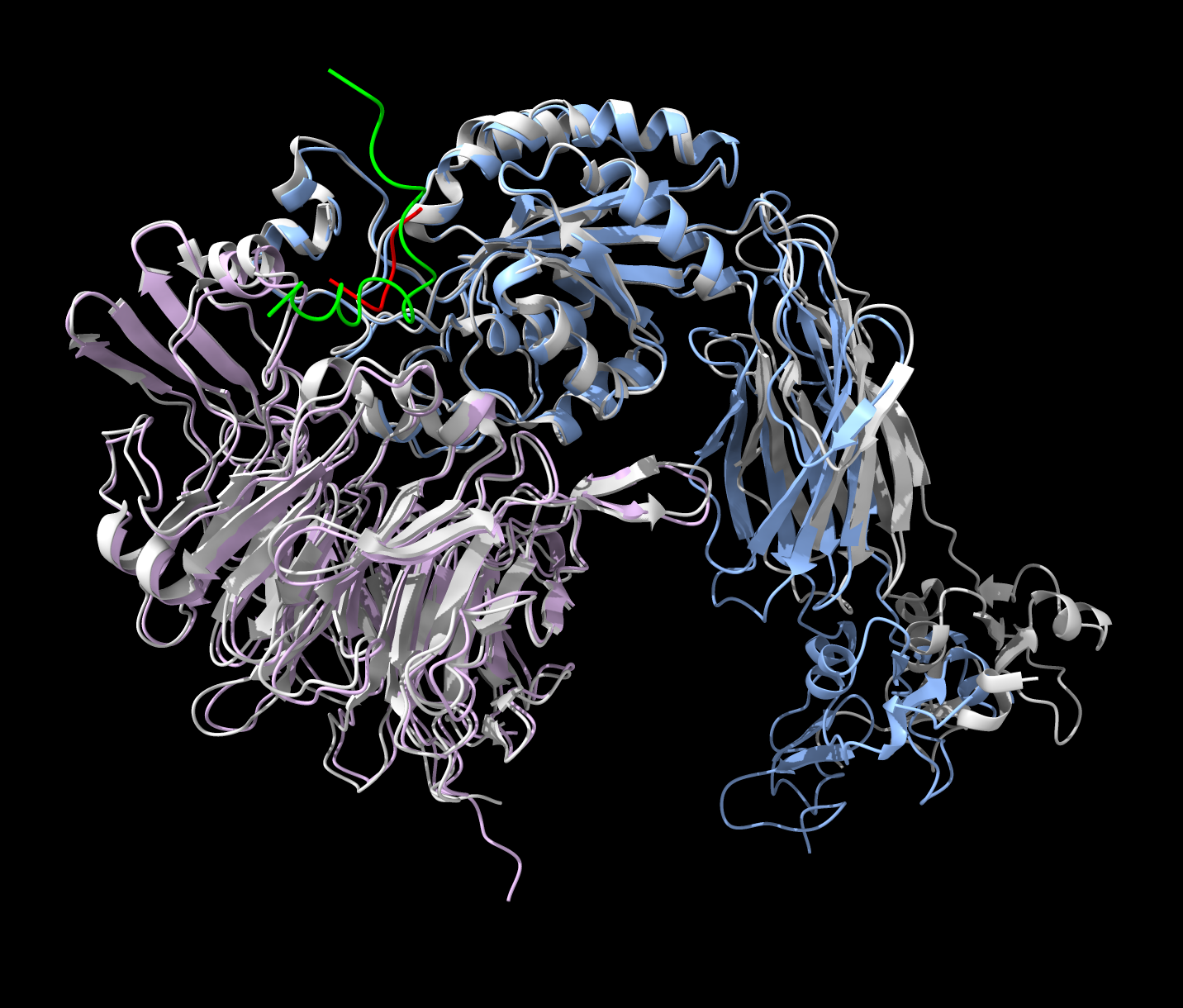
Fibrinogen + Integrin dimer predictions for Marc Schuman
Marc interested in stopping plaque formation in blood vessels by manipulating fibrinogen and integrin binding.
Ran AlphaFold prediction of human integrin alphaIIb-beta3 (pink and blue) and C-terminal 20 amino acids of fibrin gamma chain (green) superimposed on experimental structure PDB 3zdy (gray) with an RGD binding peptide (red). AlphaFold gets the right location for the peptide (green).
AlphaFold protein-protein interaction prediction tutorial for John Gross lab
Tutorial at John Gross group meeting. 4 other labs attended.
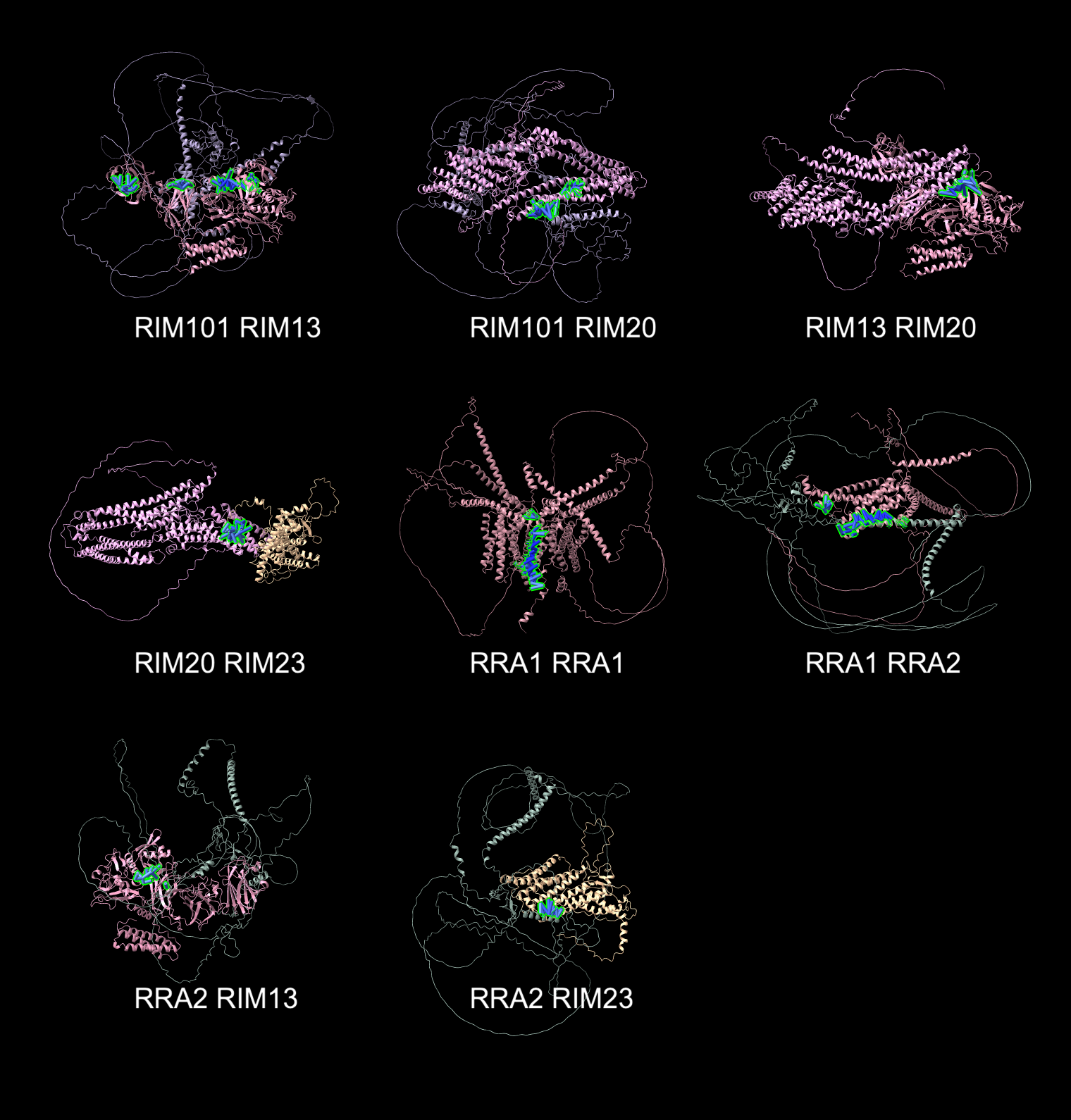
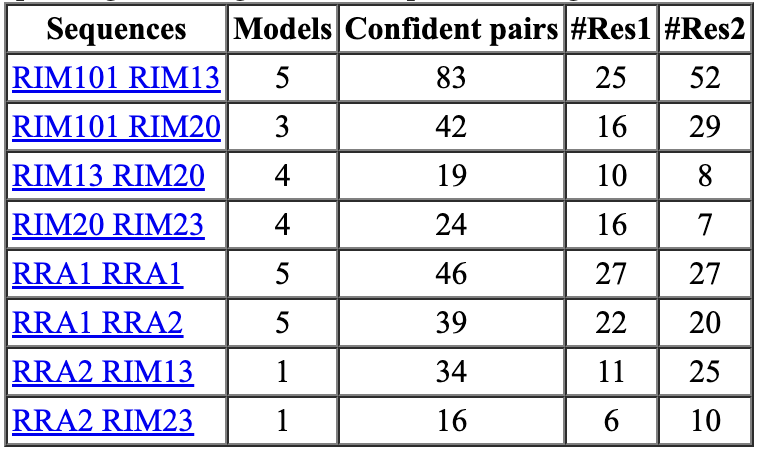
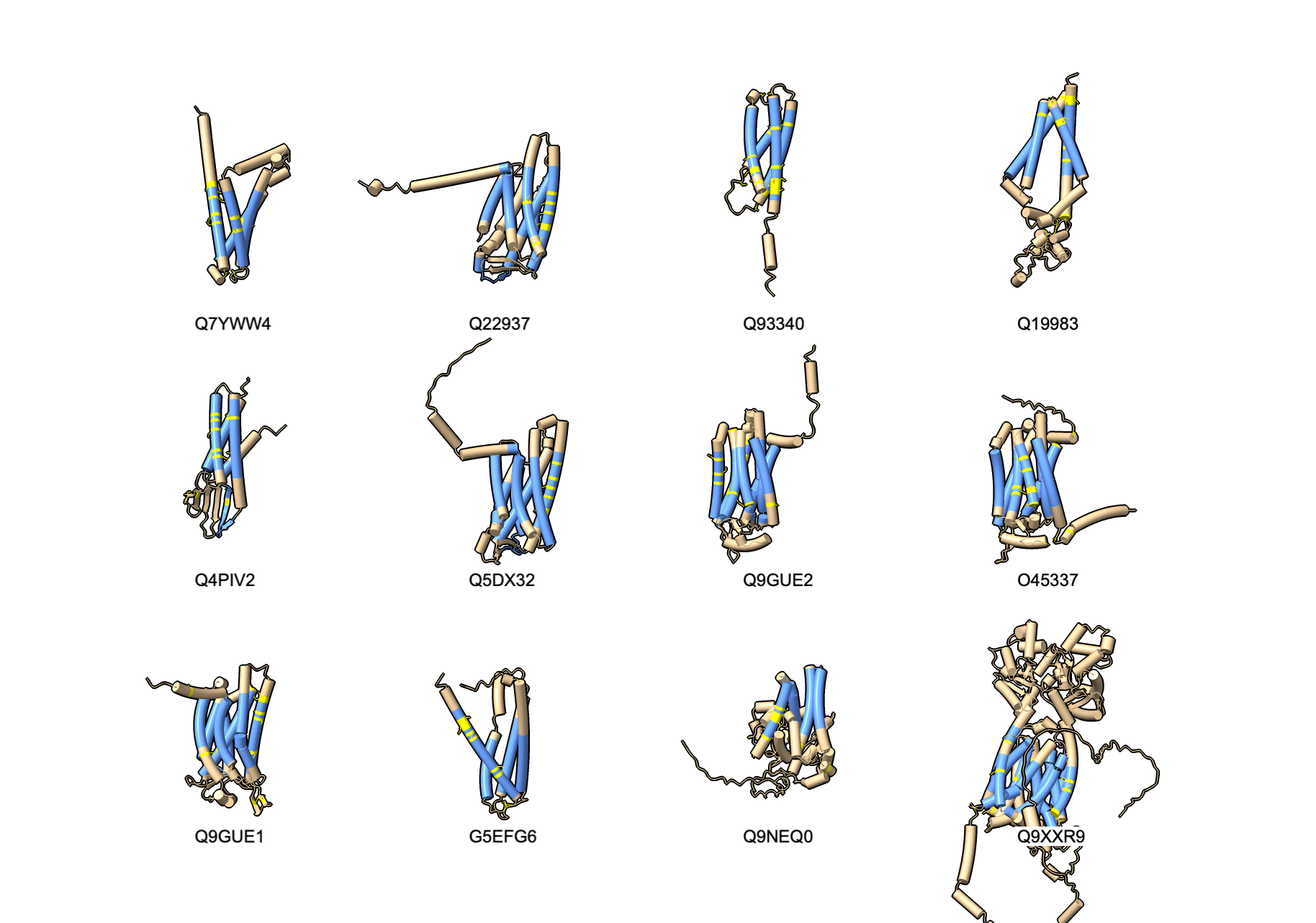
Structure Search in AlphaFold Database with Dengke Ma
Search AlphaFold database for transmembrane structures with lots of cysteines in
specific spatial arrangements for Dengke Ma who is studying such a protein that does electron transport.
Found 53 of 19827 AlphaFold database C elegans predictions satisfying
- entries with 4 or more annotated transmembrane helices, among which -
- with at least one cysteine pair (CC, CxC, CxxC) in at least two transmembrane helices, among which -
- with close (<10A) distance for cysteine pairs from two transmembrane helices.
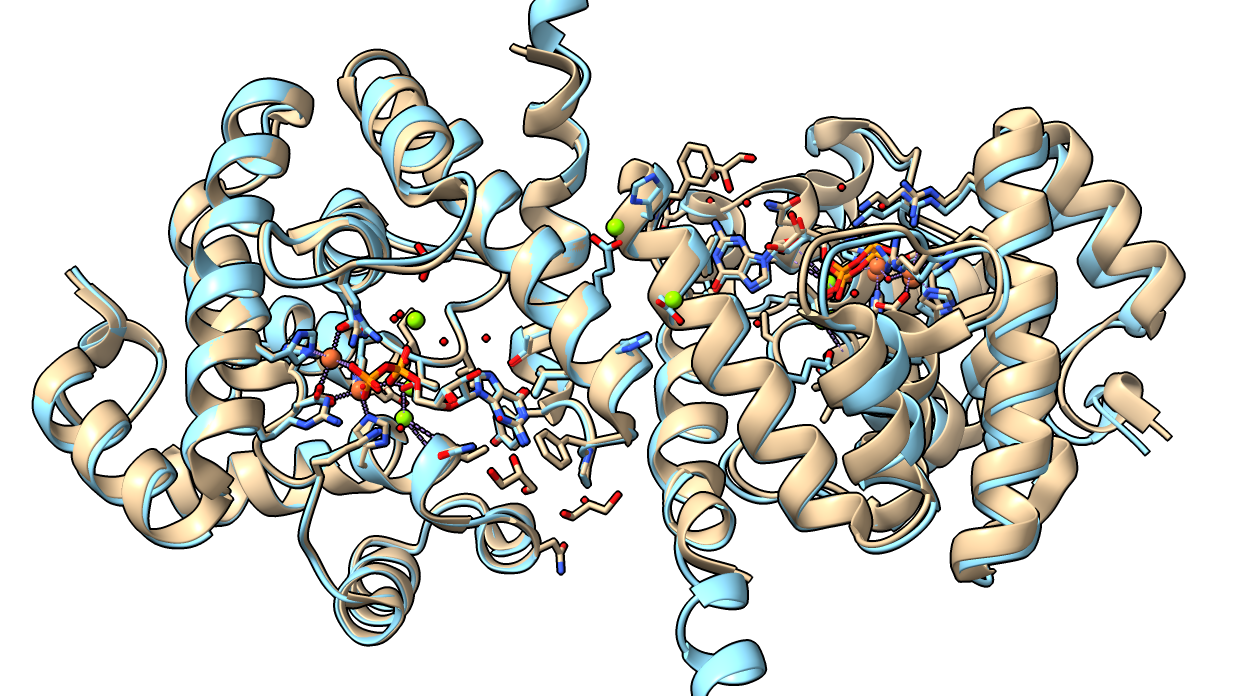
AlphaFold 3
- AlphaFold 3 published last week.
- Predicts complexes of proteins, nucleic acids, ligands, modified residues (e.g. glycosylation), ions.
- Fast and more accurate than AlphaFold 2. I ran 860 residue dimer (PDB 8jk9) with GDP ligand and Fe and Mg ions, took 2 minutes (image at right, tan 8jk9, blue AF3) and correct except for Mg ions.
- Only available through Google's AlphaFold Server web page.
- Server only allows predictions with a few common ligands like ATP.
- Google has started a drug discovery company Isomorphic Labs to utilize AlphaFold 3.
- Outcry for not releasing code. Stephanie Wankowicz authored letter with Jaime Fraser and Brian Schoichet and 1000 cosigners about code not being released. To be submitted as a Nature Letter. Pushmeet Kohli, VP of Research at Google DeepMind, said on Twitter responded that AlphaFold 3 will be released in 6 months.
- Nature editorial talks about why they violated their own rules about releasing code to publish AlphaFold 3.
- Update ChimeraX to handle AlphaFold predicted aligned error which includes PAE values for individual ligand atoms.
AlphaFold 3 Example
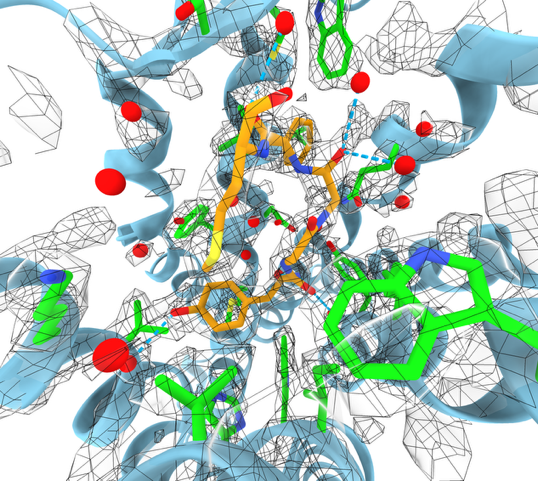
Try running AlphaFold 3 on PDB 8vdn DNA Ligase 1 protein bound to double stranded nicked DNA and AMP ligand.
Prediction took about 5 minutes. 658 protein residues, 36 DNA residues, 1 AMP molecule.
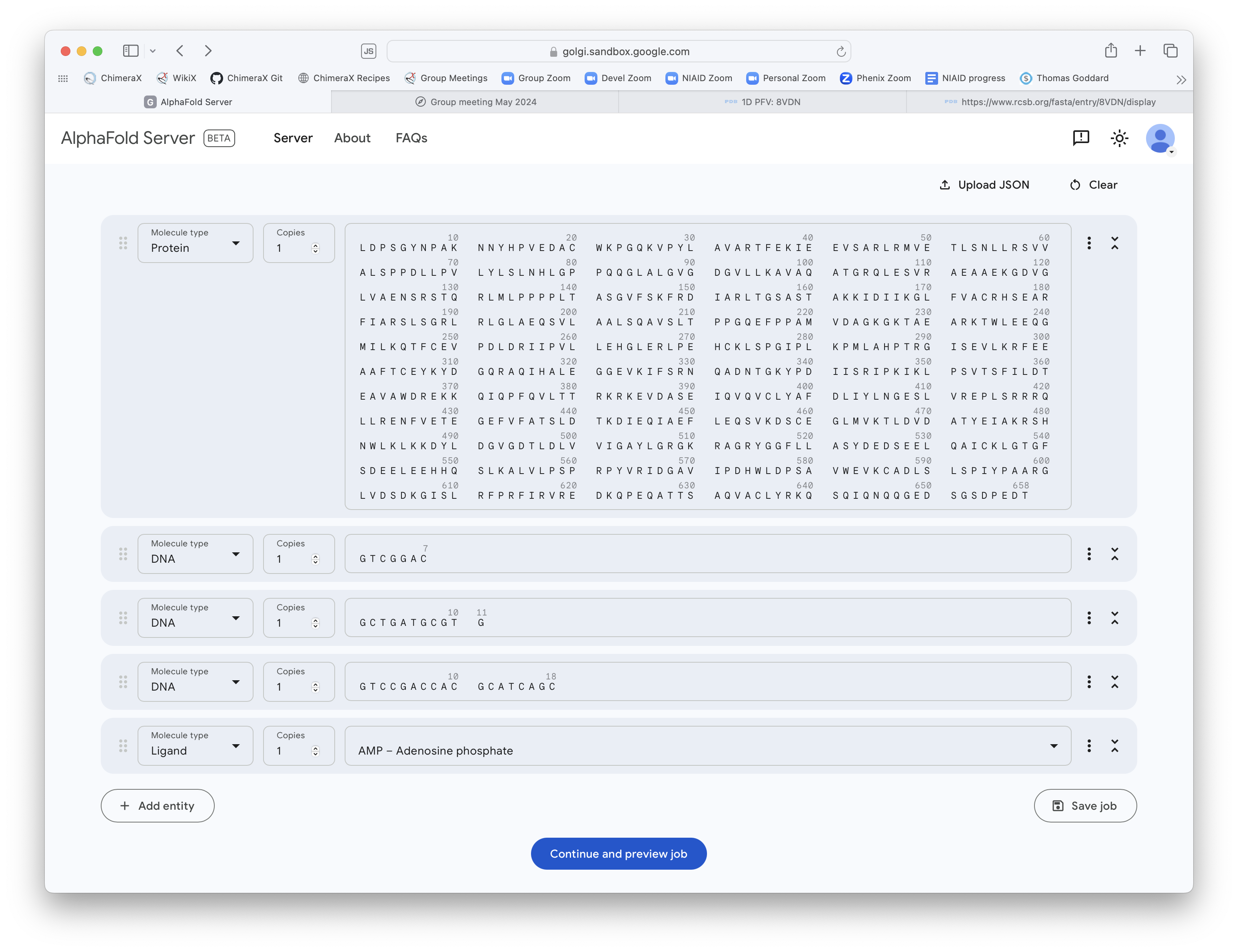
AlphaFold 3 web page input
| 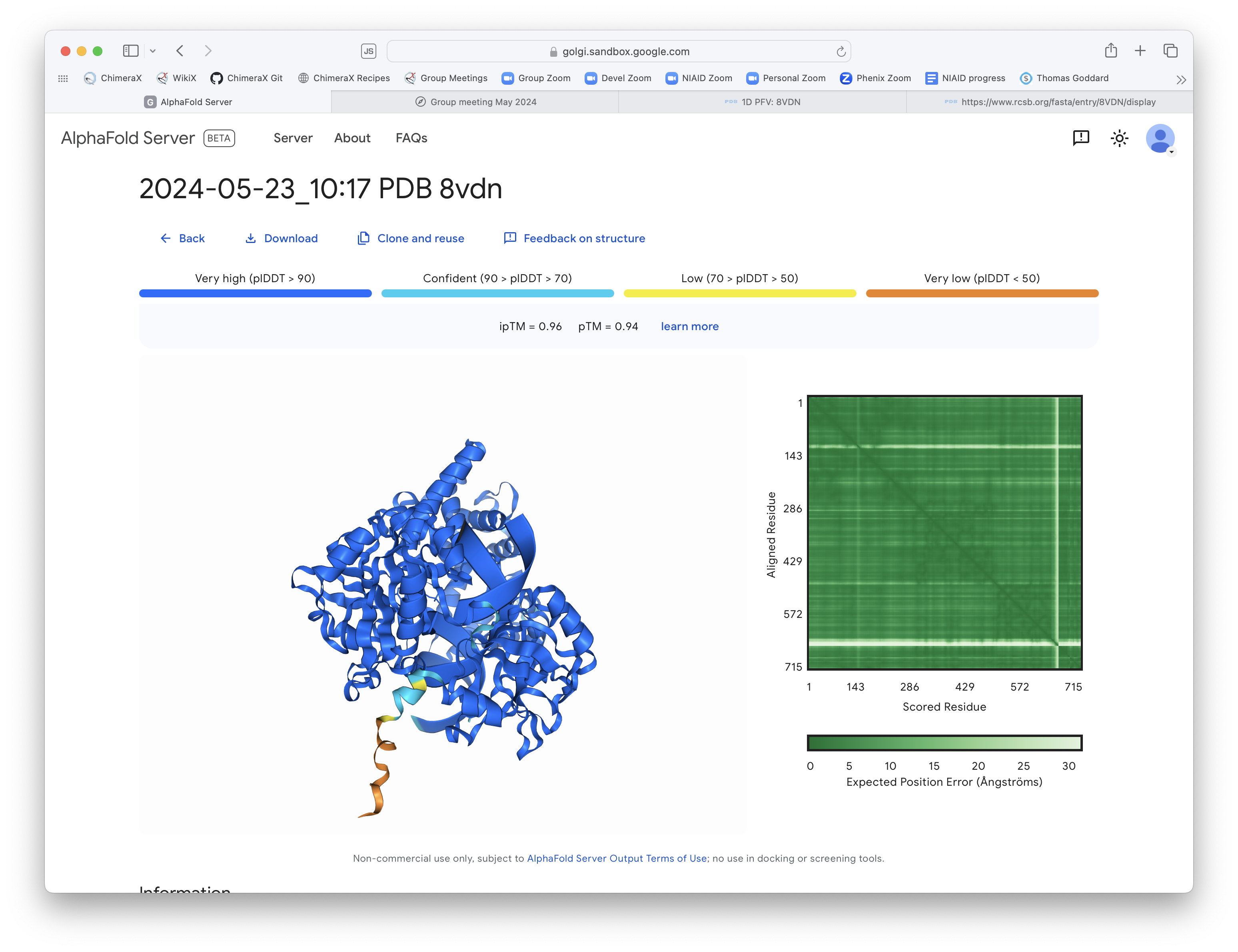
AlphaFold 3 web page output
|
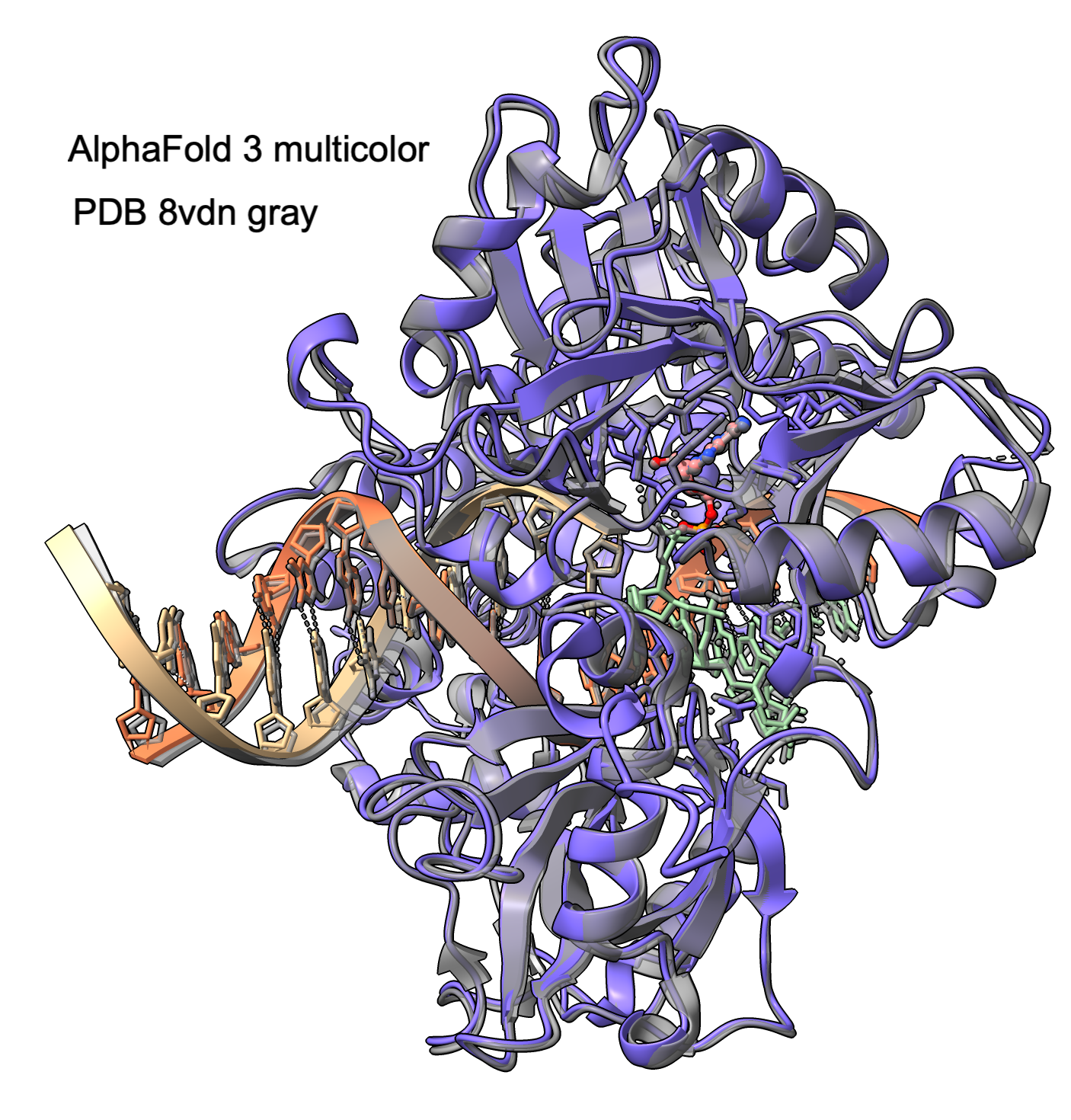
AF3 prediction and PDB (gray) superimposed in ChimeraX.
|
| 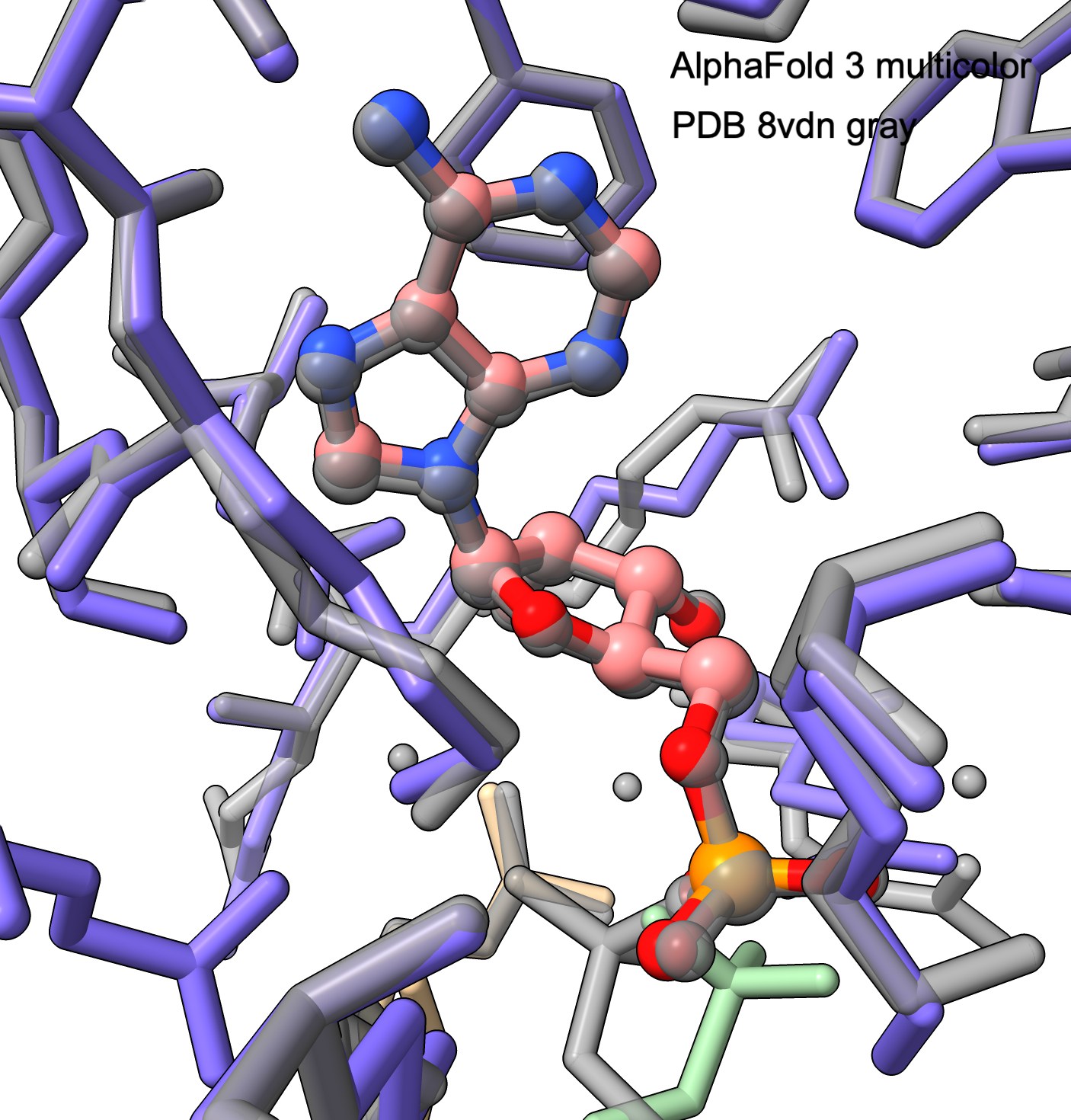
AMP ligand AF3 prediction and PDB (gray) superimposed.
|
New ChimeraX cryoEM fitting tools
| 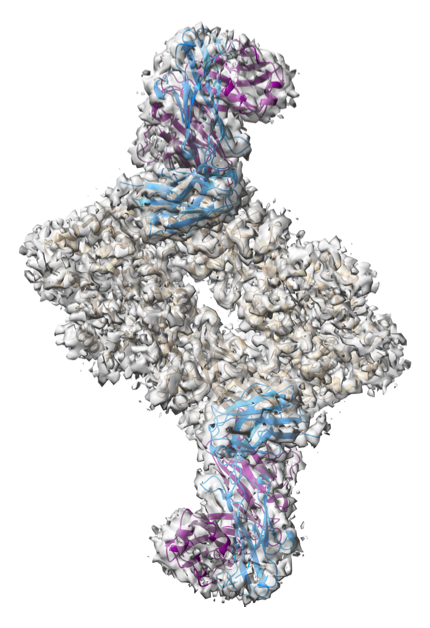
| 
|
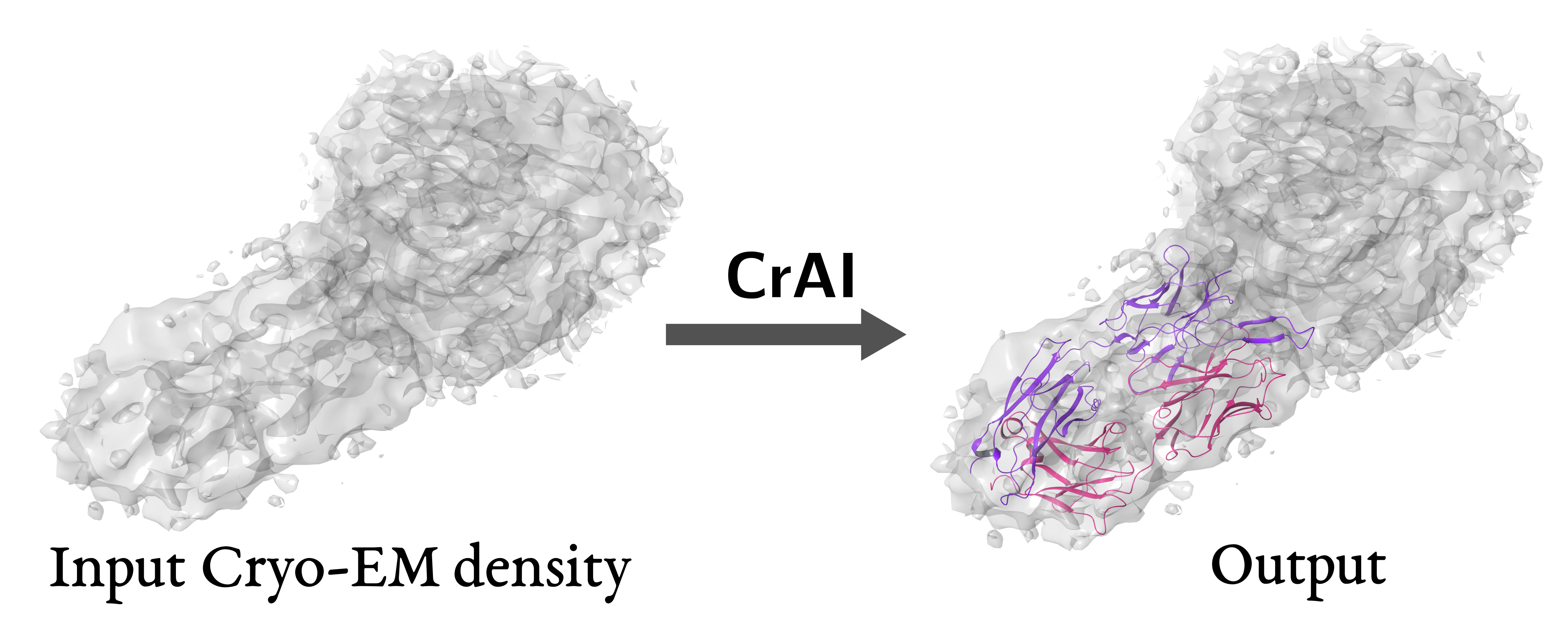
- Tested new ChimeraX Toolshed bundle CrAI that uses machine learning method to locate antibodies in cryoEM maps
and suggested improvements and tweeted (image at right).
|
- Helped Roden Luo develop DiffFit ChimeraX plugin for GPU accelerated fitting in cryoEM maps.
|
- Reviewed Randy Read's EM fitting manuscript that compares his method to ChimeraX fitmap global search.
|
Worm ribbon style graphics
- Added worm ribbon rendering code to ChimeraX, will be in 1.8 release.
- Eric made C++ data structure changes and Render by Attribute worm gui.
Virtual Reality Improvements and Demonstrations
- Added hand-gesture input to LookSee (instead of controllers in hands). Now using hand-gestures and pass-through video for all VR demos. This is most comfortable for newcomers to VR.
- Added pass-through video for ChimeraX PC VR, an NIAID request. Also added toolbar button to toggle pass-through.
- Added hand-controller "button lock" mode so demos can avoid accidentally moving models when handing off controllers, an NIAID request.
- Helped Peter Werba setting up 10 Quest VR headsets for Allison William's lab.
VR Demonstrations
- Demoed looksee with hand tracking for 15 applicants to UCSF Pharmaceutical Sciences and Pharmacogenomics (PSPG) program March 1.
- VR demo after Aashish Manglik's Byer's Lecture, showed opioid receptor and ligands and cryoEM to a dozen visitors (image at right). Discussed with Dave Agard soon to release Samsung auto-stereo displays for viewing electron tomograms.
- Gave paxlovid LookSee with hand-gestures demo at NIAID workshop.
ColabFold on Wynton
- Setup ColabFold on Wynton February 2024.
- Runs 2-10x faster than AlphaFold depending on sequence length. Some Wynton ColabFold timings here.
- Handled up to 4700 residue complexes with Nvidia A40 GPU with 48 GB.
- Need to run in two steps, first sequence alignment which needs internet access, then GPU structure calculation.
- Made tutorial for other Wynton users.
- Helped various UCSF researchers use it: Adrian Pelin in Nevan Krogan lab, ...
- Tested multiple-sequence alignment server on Plato used for running ColabFold without relying on Korea server. Will need 1 Tbyte memory to hold sequence database indices in memory.
Speeding up AlphaFold predictions without Nvidia GPU
- JAX Nvidia GPU acceleration used by AlphaFold to run structure predictions.
- Ran jax "cost analysis" on 444 amino acid dimer PDB 8rv4, took 607e9 total float32 operations.
- Mac Studio M2 single core C code float32 speed test gave 40 Gflops on CPU only.
- Might be able to run structure predictions very fast on CPU -- but cost analysis may be wrong.
- Mac also has much more compute speed on GPU and Neural Cores but those require Mac-specific APIs.

Miscellaneous
- Cover image of Tools for Protein Science 2024 used AlphaFold insulin receptor images from our ChimeraX article.
- Coauthor on integrative hybrid modeling paper IHMCIF: An Extension of the PDBx/mmCIF Data Standard for Integrative Structure Determination Methods.
- Letter of support to help Hiten Madhani (UCSF) use AlphaFold for R01 grant "Exploiting a high-precision phenotypic map to illuminate environmental sensing by a fungal meningitis pathogen."
- Letter of support for David Fay (U. Wyoming) to use AlphaFold for kinase-substrate interactions and develop a beginners ChimeraX and AlphaFold guide.
- ChimeraX 1.8 release candidates started, aiming for June 1, 2024 release.
Vacation in China
Spent 3 weeks in China visiting family in cities Kunming, Guiyan and Chongqing in the south.
Work from China
- Fixed ChimeraX AlphaFold predictions from China via iPhone screensharing to work! Ugh. Google Colab changed Python notebook web page so sequence sending broke. Fix required new ChimeraX.
- Last time Google Colab broke ChimeraX AlphaFold was Christmas day 2023.