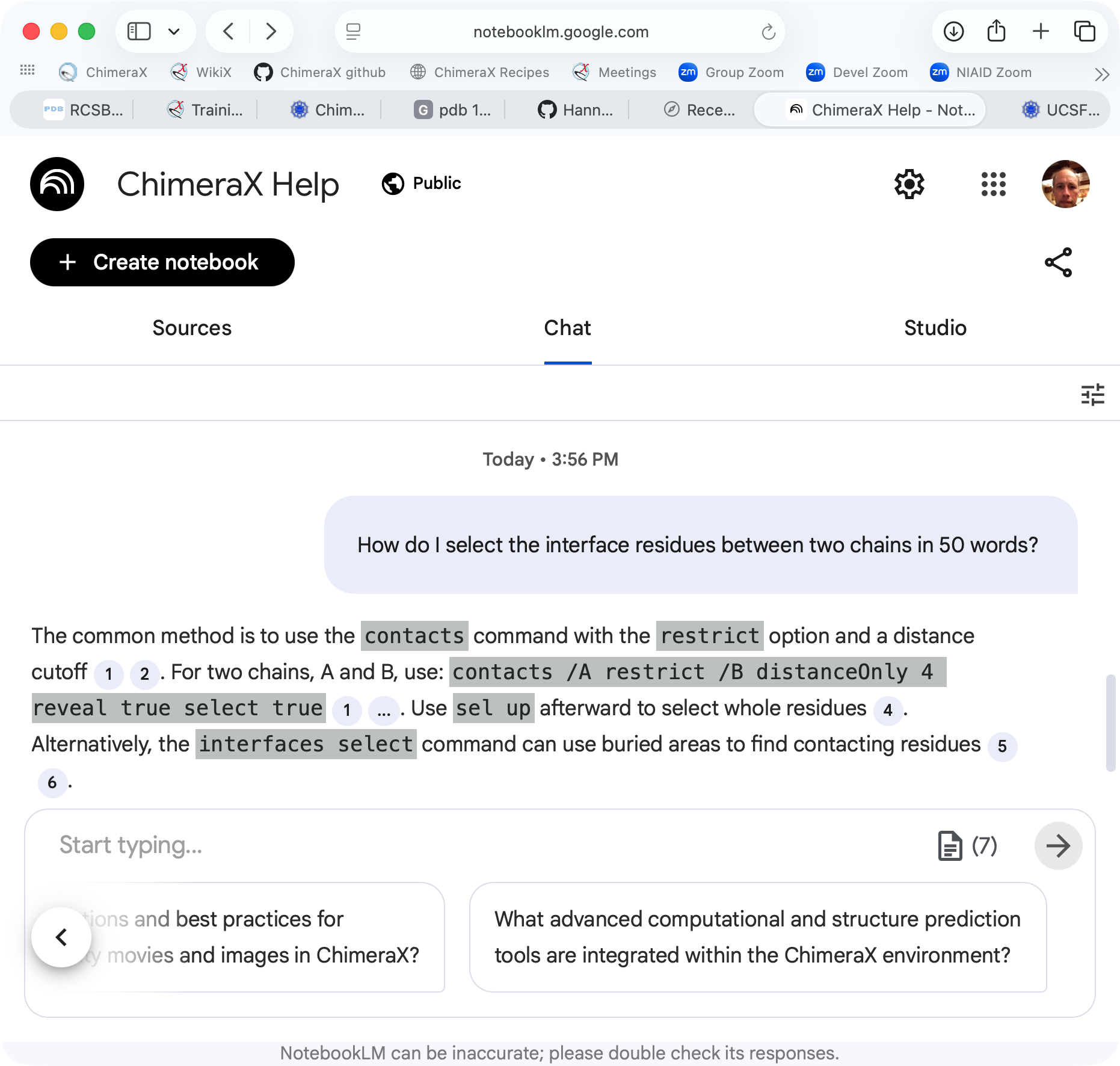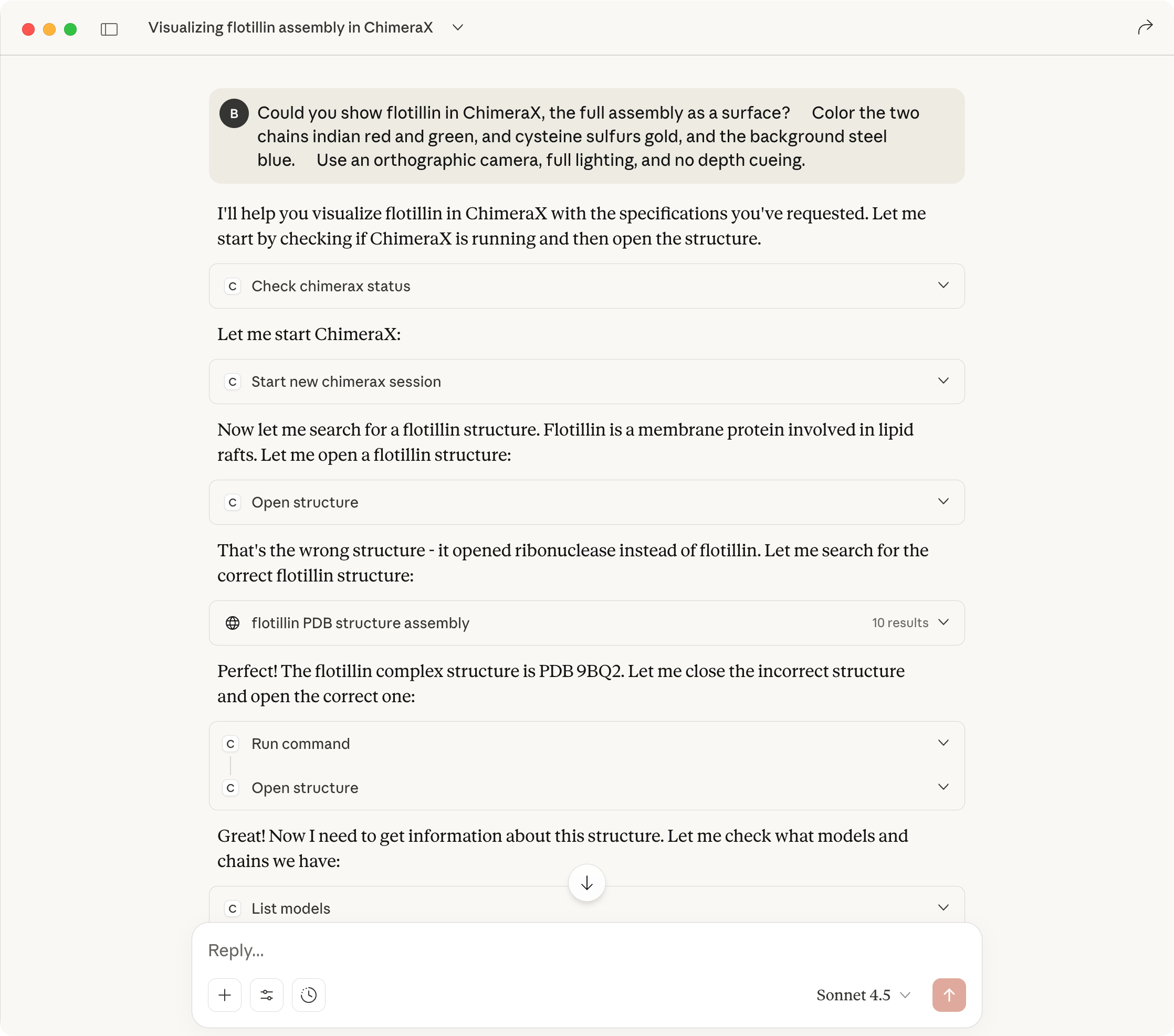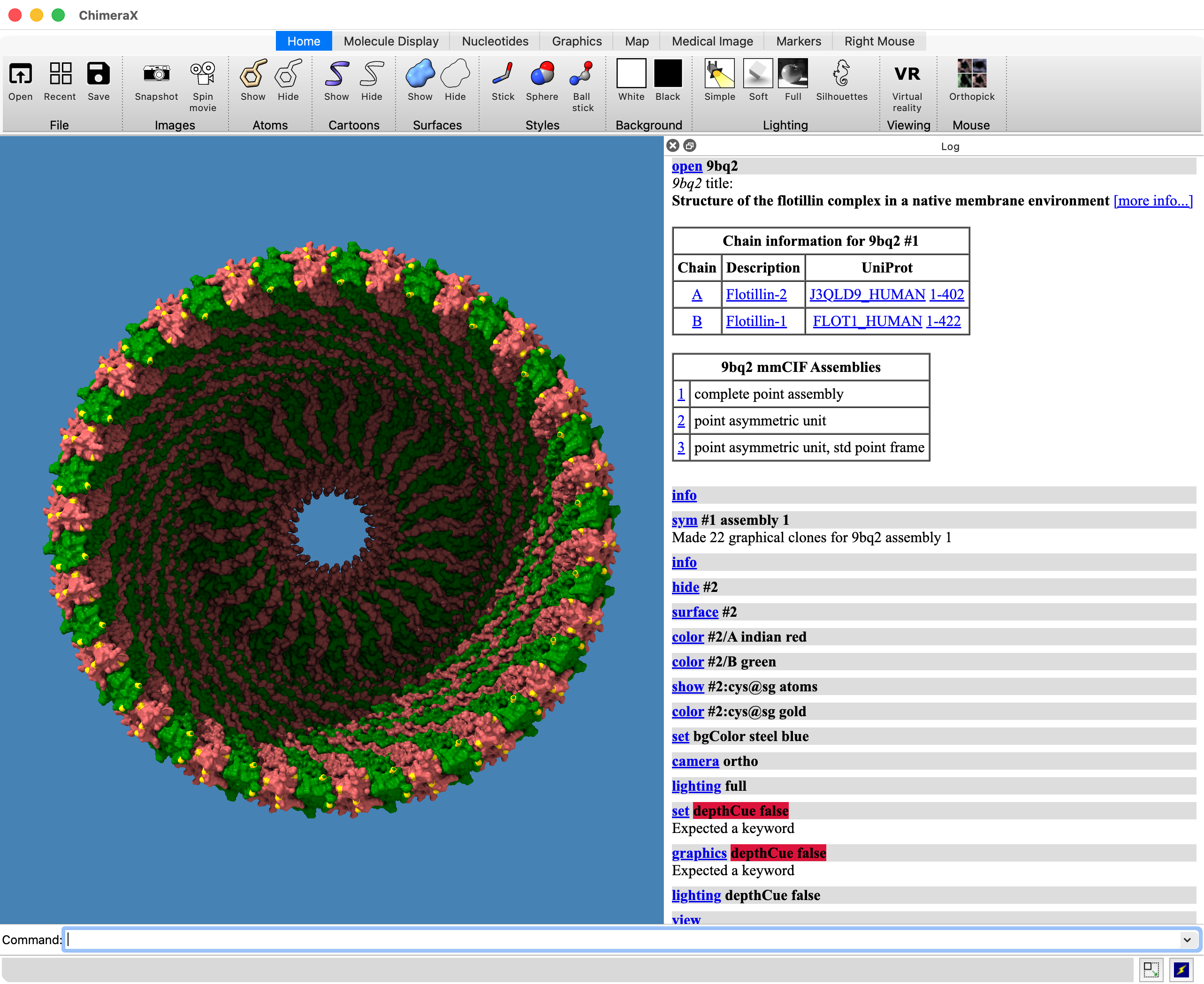
10 BoltzGen designed proteins (blue to red) binding GM2 activator (tan)
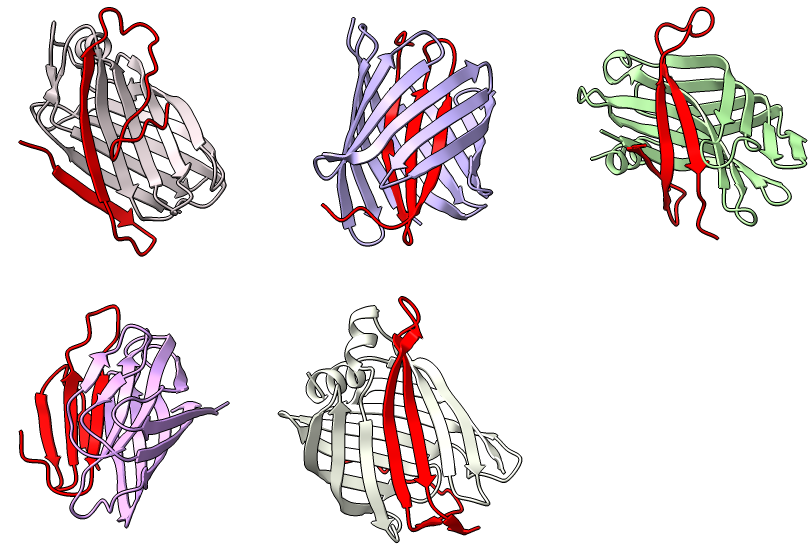
5 BoltzGen designed proteins binding amyloid beta (red)
design_to_target_iptm 0.63 0.57 0.51 0.46 0.41 ...
min_design_to_target_pae 2.9 3.1 4.4 5.3 5.2 ...
interaction_pae 10.3 10.2 14.0 15.2 16.3 ...
design_ptm 0.81 0.78 0.86 0.82 0.79 ...
num_prot_tokens 256 244 291 296 296 ...
...
design_to_target_iptm 0.62 0.64 0.60 0.60 0.54
min_design_to_target_pae 1.2 2.2 2.1 2.0 2.6
interaction_pae 10.5 8.8 10.5 11.8 11.6
design_ptm 0.77 0.77 0.74 0.72 0.72
num_prot_tokens 168 176 176 162 180
...