December 25, 2025
The RBVI wishes you a safe and happy holiday season!
See our
2025 card and the
gallery of previous cards back to 1985.
December 16, 2025
The ChimeraX 1.11 production release is
available! See the
change log
for what's new.
November 21, 2025
The ChimeraX 1.11 release candidate is
available –
please try it and report
any issues. See the
change log
for what's new.
This will be the last release to support Red Hat Enterprise Linux 8 and
its derivatives.
Previous news...
UCSF ChimeraX
UCSF ChimeraX (or simply ChimeraX)
is the next-generation molecular visualization program from the
Resource for Biocomputing,
Visualization, and Informatics (RBVI),
following UCSF Chimera.
ChimeraX can be downloaded free of charge
for academic, government, nonprofit, and personal use.
Commercial users, please see
ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
 ChimeraX on Bluesky:
@chimerax.ucsf.edu
ChimeraX on Bluesky:
@chimerax.ucsf.edu
Different representations of nucleotides can be shown with the
nucleotides
command or Toolbar
icons. Options include filled rings, slabs for bases
(box, muffler, or ellipsoid shape), bumps on slabs to show base orientation,
simple tubes instead of ribose atoms, and continuous or broken ladder rungs.
Nucleotide representations can be the same color as the ribbon
or a different color, and multiple nucleotide styles can be used
within a single structure.
See also: Presets menu
More features...
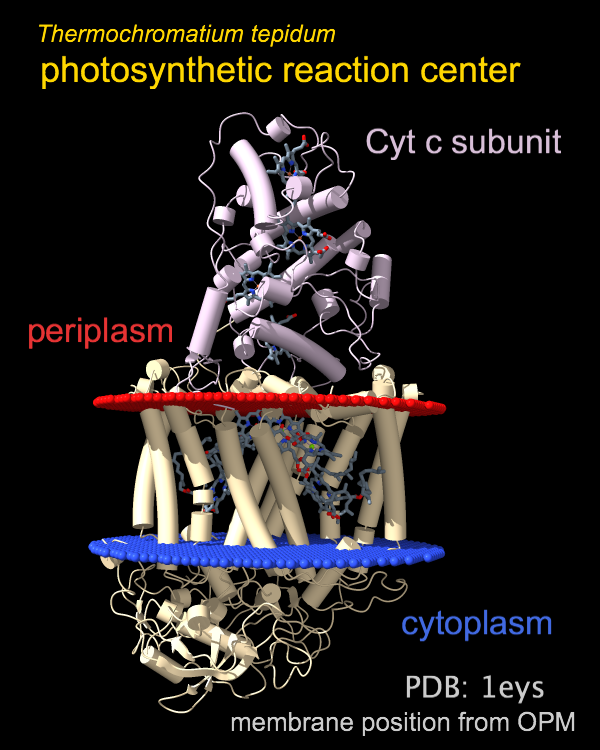
The photosynthetic reaction center from a
purple sulfur bacterium is shown as a cartoon with “tube” helices
and membrane boundaries from the OPM database (Orientations of Proteins in Membranes,
entry 1eys).
Blue and red balls represent the cytoplasmic and periplasmic sides
of the bacterial inner membrane, respectively.
The title and other text labels were added with the
2dlabels
command and repositioned interactively with the move label
mouse mode
 .
ChimeraX session file: prc.cxs
.
ChimeraX session file: prc.cxs
More images...