December 25, 2025
The RBVI wishes you a safe and happy holiday season!
See our
2025 card and the
gallery of previous cards back to 1985.
December 16, 2025
The ChimeraX 1.11 production release is
available! See the
change log
for what's new.
November 21, 2025
The ChimeraX 1.11 release candidate is
available –
please try it and report
any issues. See the
change log
for what's new.
This will be the last release to support Red Hat Enterprise Linux 8 and
its derivatives.
Previous news...
UCSF ChimeraX
UCSF ChimeraX (or simply ChimeraX)
is the next-generation molecular visualization program from the
Resource for Biocomputing,
Visualization, and Informatics (RBVI),
following UCSF Chimera.
ChimeraX can be downloaded free of charge
for academic, government, nonprofit, and personal use.
Commercial users, please see
ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
 ChimeraX on Bluesky:
@chimerax.ucsf.edu
ChimeraX on Bluesky:
@chimerax.ucsf.edu
The 3D contents of the ChimeraX graphics window can be embedded
in a web page by:
- saving the scene
as a GLTF (*.glb) file
- referencing the GLTF file in the HTML source of the web page
(details...)
(More features...)
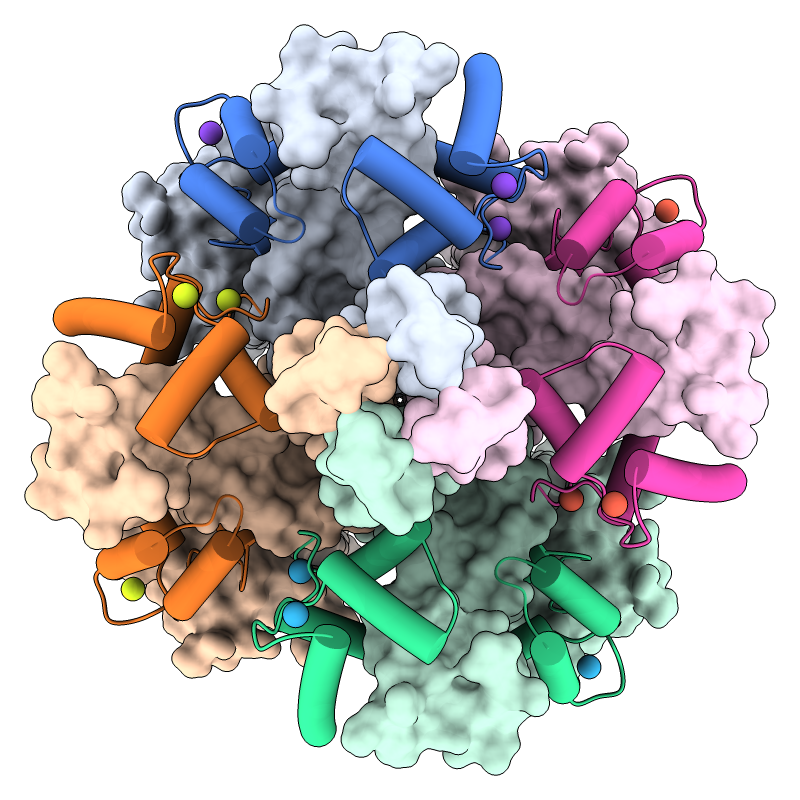
KCNQ1 is the pore-forming subunit of a cardiac potassium channel.
It binds to calmodulin, and mutations in either of these proteins
can cause congenital long QT syndrome, a dangerous
propensity for irregular heartbeats.
In the image, a structure of the KCNQ1/calmodulin complex
(PDB 5vms)
has been assembled into the native tetrameric form with the
sym command.
The view is from the cytoplasmic side, with
KCNQ1 shown as surfaces, calmodulin as cartoons, and calcium ions as balls.
A pastel palette
from ColorBrewer
has been used to color the surfaces, darkened with
color modify
for the cartoons, and “rotated” 45° in hue for the ions.
See the command file colormod.cxc.
More images...