

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Recent Citations
Tubulin code eraser CCP5 binds branch glutamates by substrate deformation. Chen J, Zehr EA et al. Nature. 2024 Jul 25;631(8022):905–912.
A μ-opioid receptor modulator that works cooperatively with naloxone. O'Brien ES, Rangari VA et al. Nature. 2024 Jul 18;631(8021):686–693.
Molecular mechanism of plasmid elimination by the DdmDE defense system. Loeff L, Adams DW et al. Science. 2024 Jul 12;385(6705):188-194.
Bitter taste TAS2R14 activation by intracellular tastants and cholesterol. Hu X, Ao W et al. Nature. 2024 Jul 11;631(8020):459–466.
Evolution and host-specific adaptation of Pseudomonas aeruginosa. Weimann A, Dinan AM et al. Science. 2024 Jul 5;385(6704):eadi0908.
Previously featured citations...Chimera Search
Google™ SearchNews
August 1, 2024
Planned downtime: The Chimera and ChimeraX websites, web services (Blast Protein, Modeller, ...) and cgl.ucsf.edu e-mail will be unavailable August 1, 3-6 pm PDT.
July 16, 2024
Chimera production release 1.18 is now available. See the release notes for details.
June 17-18, 2024
Planned downtime: The Chimera and ChimeraX websites, web services (Blast Protein, Modeller, ...) and cgl.ucsf.edu e-mail will be unavailable June 17-18 PDT.
Previous news...Upcoming Events

UCSF Chimera is a program for the interactive visualization and analysis of molecular structures and related data, including density maps, trajectories, and sequence alignments. It is available free of charge for noncommercial use. Commercial users, please see Chimera commercial licensing.
We encourage Chimera users to try ChimeraX for much better performance with large structures, as well as other major advantages and completely new features in addition to nearly all the capabilities of Chimera (details...).
Chimera is no longer under active development. Chimera development was supported by a grant from the National Institutes of Health (P41-GM103311) that ended in 2018.
Feature Highlight
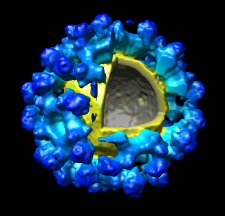
Chimera's Volume Viewer displays three-dimensional electron and light microscope data, X-ray density maps, electrostatic potential and other volumetric data. Contour surfaces, meshes and volumetric display styles are provided and thresholds can be changed interactively. Maps can be colored, sliced, segmented, and modifications can be saved. Markers can be placed and structures can be traced. The accompanying image shows a density map of Kelp fly virus from electron microscopy colored radially and with an octant cut out.
(More features...)Gallery Sample
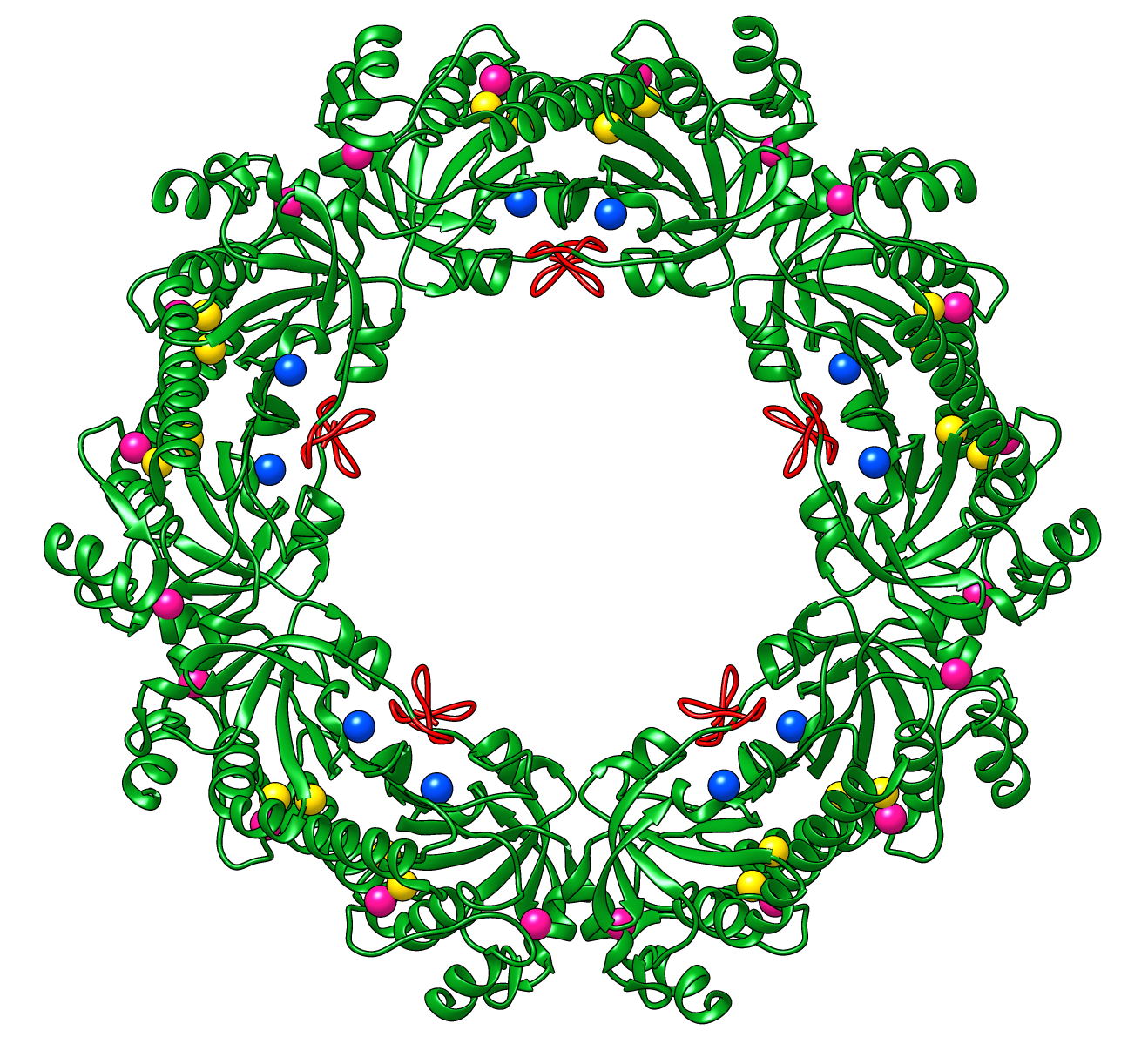
Peroxiredoxins are enzymes that help cells cope with stressors such as high levels of reactive oxygen species. The image shows a decameric peroxiredoxin from human red blood cells (Protein Data Bank entry 1qmv), styled as a holiday wreath.
See also the RBVI holiday card gallery.
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.