

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Recent Citations
Structural insights into the activation mechanism of the human metabolite receptor HCAR1. Gao M, Zang S et al. Sci Signal. 2026 Jan 6;19(919):eadw1483.
Crystal structure of Methanococcus jannaschii dihydroorotase with substrate bound. Vitali J, Nix JC et al. Acta Crystallogr F Struct Biol Commun. 2026 Jan 1;82(Pt 1):23-31.
Correlation between solvation free energy and solute-solvent interaction energy in energy representation theory. Maruyama Y, Matubayasi N. J Phys Chem B. 2025 Dec 25;129(51):13230-13241.
Structural snapshots capture nucleotide release at the μ-opioid receptor. Khan S, Tyson AS et al. Nature. 2025 Dec 18;648(8094):755–763.
Bottom-up design of Ca2+ channels from defined selectivity filter geometry. Liu Y, Weidle C et al. Nature. 2025 Dec 11;648(8093):468–476.
Previously featured citations...Chimera Search
Google™ SearchNews
December 25, 2025

|
September 22, 2025
Mac users may wish to defer upgrading to MacOS Tahoe. Currently on that OS the Chimera graphics window is shifted so that it covers the command and status lines.
March 6, 2025
Chimera production release 1.19 is now available, fixing the ability to fetch structures from the PDB (details...).
Previous news...Upcoming Events

UCSF Chimera is a program for the interactive visualization and analysis of molecular structures and related data, including density maps, trajectories, and sequence alignments. It is available free of charge for noncommercial use. Commercial users, please see Chimera commercial licensing.
We encourage Chimera users to try ChimeraX for much better performance with large structures, as well as other major advantages and completely new features in addition to nearly all the capabilities of Chimera (details...).
Chimera is no longer under active development. Chimera development was supported by a grant from the National Institutes of Health (P41-GM103311) that ended in 2018.
Feature Highlight
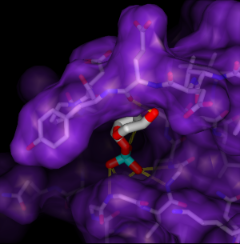
Given the structures of ligand and receptor molecules, docking programs calculate possible binding modes. In virtual screening, small organic compounds (typically from a database of many thousands) are treated as possible ligands, and a target macromolecule is treated as the receptor.
ViewDock facilitates the interactive selection of compounds from the output of docking programs, including DOCK and Glide. The hits can be viewed in the context of the binding site and sorted or screened by various properties such as score or number of hydrogen bonds to the receptor. The Dock Prep tool can be used to prepare structures for docking or other calculations by adding hydrogens, assigning partial charges, and performing other related tasks.
(More features...)
Gallery Sample
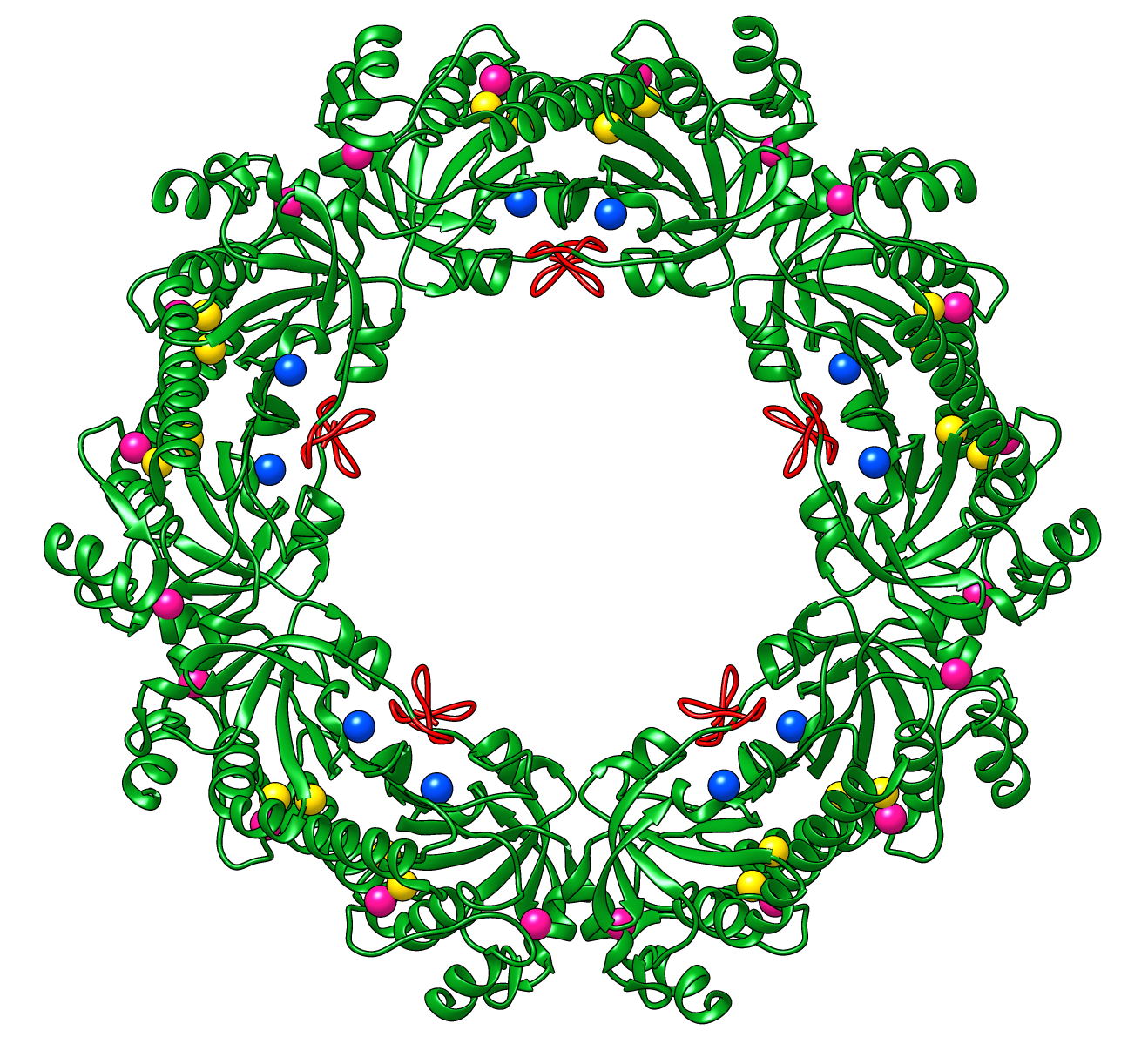
Peroxiredoxins are enzymes that help cells cope with stressors such as high levels of reactive oxygen species. The image shows a decameric peroxiredoxin from human red blood cells (Protein Data Bank entry 1qmv), styled as a holiday wreath.
See also the RBVI holiday card gallery.
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.