

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
Plug-in strategy for resistance engineering inspired by potato NLRome. Wang L, Li H et al. Nature. 2026 Jan 8;649(8096):396–405.
Deep contrastive learning enables genome-wide virtual screening. Jia Y, Gao B et al. Science. 2026 Jan 8;391(6781):eads9530.
Recurrent acquisition of nuclease-protease pairs in antiviral immunity. Tuck OT, Hu JJ et al. Science. 2026 Jan 8;391(6781):195-201.
Asynchronous subunit transitions prime acetylcholine receptor activation. Thompson MJ, Tessier CJG et al. Science. 2026 Jan 1;391(6780):eadw1264.
Mechanism of cotranslational modification of histones H2A and H4 by MetAP1 and NatD. Yudin D, Jaskolowski M et al. Sci Adv. 2025 Dec 19;11(51):eaeb1017.
More citations...News
December 25, 2025

|
December 16, 2025
The ChimeraX 1.11 production release is available! See the change log for what's new.
November 21, 2025
The ChimeraX 1.11 release candidate is available – please try it and report any issues. See the change log for what's new. This will be the last release to support Red Hat Enterprise Linux 8 and its derivatives.
Previous news...Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
 ChimeraX on Bluesky:
@chimerax.ucsf.edu
ChimeraX on Bluesky:
@chimerax.ucsf.edu
Feature Highlight
 |
 |
 |
 |
 |
 |
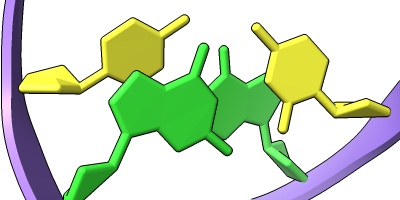
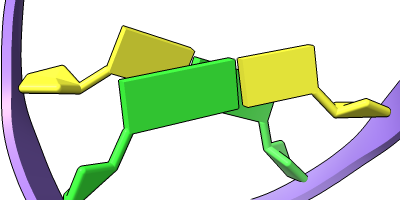
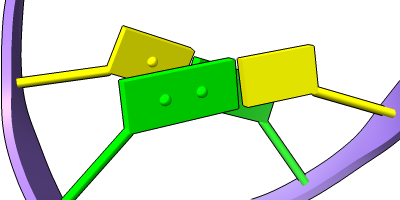
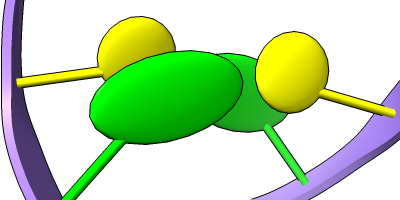
Different representations of nucleotides can be shown with the nucleotides command or Toolbar icons. Options include filled rings, slabs for bases (box, muffler, or ellipsoid shape), bumps on slabs to show base orientation, simple tubes instead of ribose atoms, and continuous or broken ladder rungs. Nucleotide representations can be the same color as the ribbon or a different color, and multiple nucleotide styles can be used within a single structure.
See also: Presets menu
More features...
Example Image
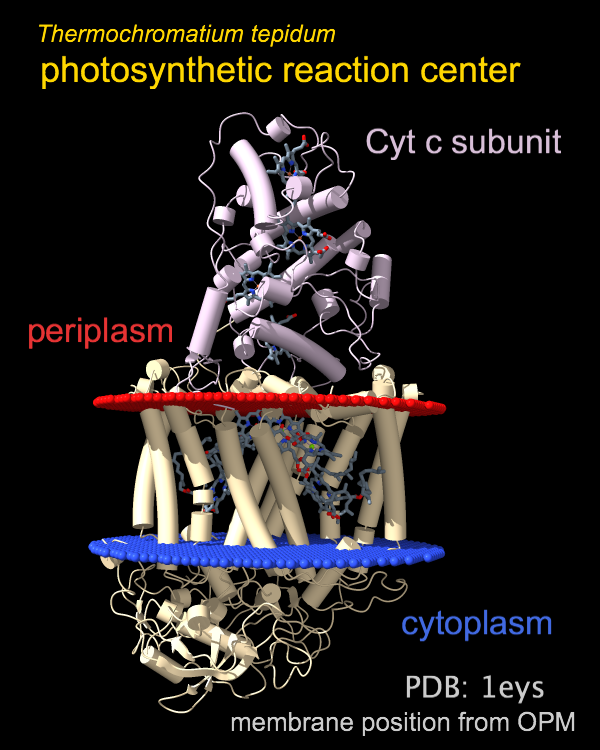
The photosynthetic reaction center from a
purple sulfur bacterium is shown as a cartoon with “tube” helices
and membrane boundaries from the OPM database (Orientations of Proteins in Membranes,
entry 1eys).
Blue and red balls represent the cytoplasmic and periplasmic sides
of the bacterial inner membrane, respectively.
The title and other text labels were added with the
2dlabels
command and repositioned interactively with the move label
mouse mode
![]() .
ChimeraX session file: prc.cxs
.
ChimeraX session file: prc.cxs
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.